Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN-117 [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN-117 Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 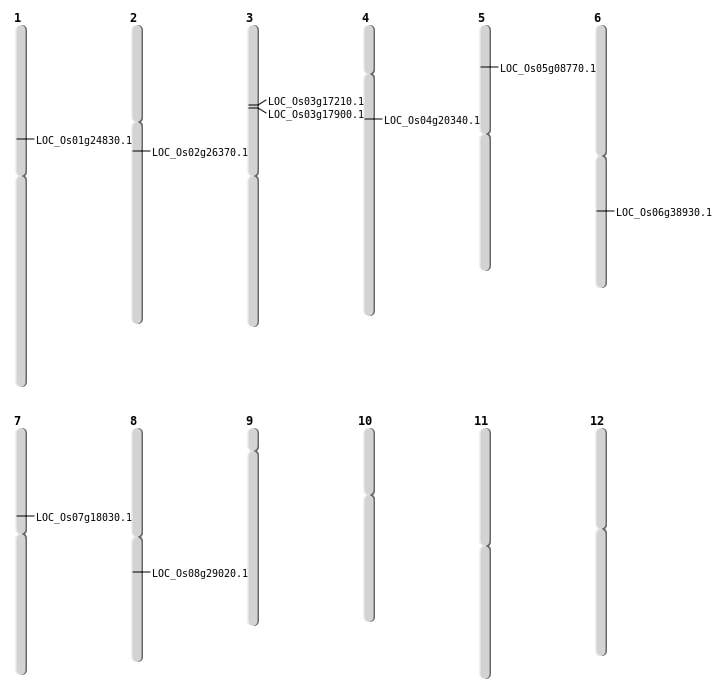
|
| Variant Information |
|---|
Single base substitutions: 21
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 12748987 | T-A | HET | INTERGENIC | |
| SBS | Chr1 | 13985196 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os01g24830 |
| SBS | Chr1 | 17847249 | A-T | HET | INTRON | |
| SBS | Chr1 | 40472121 | C-A | HOMO | INTERGENIC | |
| SBS | Chr10 | 21101407 | C-T | HOMO | INTERGENIC | |
| SBS | Chr11 | 15371721 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 5691925 | G-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 20147117 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 33168769 | A-T | HET | INTRON | |
| SBS | Chr2 | 33168770 | A-T | HET | INTRON | |
| SBS | Chr3 | 31900522 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 10429411 | G-C | HET | INTERGENIC | |
| SBS | Chr4 | 14642560 | G-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 27261419 | C-T | HOMO | INTRON | |
| SBS | Chr5 | 10430129 | G-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr5 | 5799478 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr6 | 21172617 | A-C | HET | INTERGENIC | |
| SBS | Chr7 | 10674140 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os07g18030 |
| SBS | Chr7 | 21219739 | C-A | HOMO | INTERGENIC | |
| SBS | Chr8 | 7853624 | C-A | HET | INTERGENIC | |
| SBS | Chr9 | 11639168 | C-G | HET | INTERGENIC |
Deletions: 19
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr9 | 6983199 | 6983210 | 11 | |
| Deletion | Chr3 | 9972649 | 9972995 | 346 | LOC_Os03g17900 |
| Deletion | Chr12 | 10703131 | 10703134 | 3 | |
| Deletion | Chr4 | 11382858 | 11382859 | 1 | LOC_Os04g20340 |
| Deletion | Chr9 | 11959828 | 11959832 | 4 | |
| Deletion | Chr11 | 14565397 | 14565599 | 202 | |
| Deletion | Chr8 | 17767328 | 17767337 | 9 | LOC_Os08g29020 |
| Deletion | Chr6 | 18089306 | 18089309 | 3 | |
| Deletion | Chr12 | 19487424 | 19487426 | 2 | |
| Deletion | Chr1 | 20656284 | 20656287 | 3 | |
| Deletion | Chr4 | 20685869 | 20685873 | 4 | |
| Deletion | Chr2 | 21381277 | 21381287 | 10 | |
| Deletion | Chr7 | 21742080 | 21742082 | 2 | |
| Deletion | Chr6 | 23087523 | 23087528 | 5 | |
| Deletion | Chr6 | 23095093 | 23095138 | 45 | LOC_Os06g38930 |
| Deletion | Chr12 | 23909752 | 23909755 | 3 | |
| Deletion | Chr2 | 24063952 | 24063965 | 13 | |
| Deletion | Chr3 | 25788077 | 25788087 | 10 | |
| Deletion | Chr4 | 28581349 | 28581408 | 59 |
Insertions: 5
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr2 | 15461328 | 15483742 | LOC_Os02g26370 |
Translocations: 3
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr5 | 4813987 | Chr3 | 9972653 | 2 |
| Translocation | Chr3 | 9585679 | Chr1 | 4749093 | |
| Translocation | Chr3 | 9593506 | Chr1 | 4749113 | LOC_Os03g17210 |
