Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN-121 [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN-121 Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 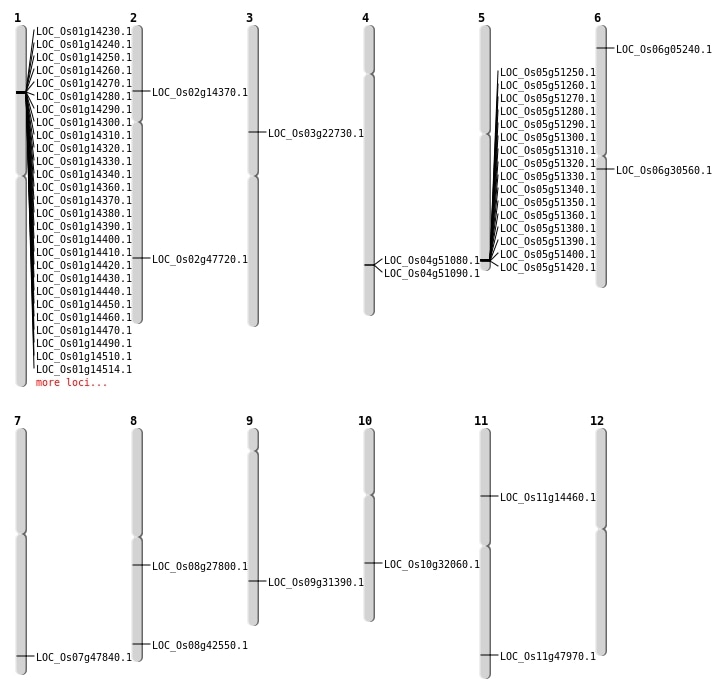
|
| Variant Information |
|---|
Single base substitutions: 33
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 13288697 | A-G | HOMO | INTERGENIC | |
| SBS | Chr1 | 41888174 | A-C | HET | INTRON | |
| SBS | Chr10 | 14080600 | C-T | HET | INTRON | |
| SBS | Chr10 | 16841876 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os10g32060 |
| SBS | Chr10 | 18668333 | C-G | HET | INTRON | |
| SBS | Chr10 | 8228602 | T-G | HET | INTERGENIC | |
| SBS | Chr11 | 18887432 | A-G | HET | INTRON | |
| SBS | Chr11 | 28934545 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os11g47970 |
| SBS | Chr11 | 28934546 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os11g47970 |
| SBS | Chr11 | 6248706 | C-A | HET | INTRON | |
| SBS | Chr11 | 6248736 | G-A | HET | INTRON | |
| SBS | Chr11 | 6871988 | T-C | HOMO | INTERGENIC | |
| SBS | Chr11 | 6871995 | T-C | HOMO | INTERGENIC | |
| SBS | Chr12 | 14391264 | G-A | HOMO | INTERGENIC | |
| SBS | Chr12 | 20810068 | C-T | HET | INTRON | |
| SBS | Chr2 | 1158391 | G-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 2686209 | T-A | HET | INTERGENIC | |
| SBS | Chr2 | 29473295 | C-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 30851755 | G-T | HET | INTERGENIC | |
| SBS | Chr2 | 32373333 | A-C | HOMO | INTRON | |
| SBS | Chr2 | 6136942 | A-G | HET | INTRON | |
| SBS | Chr2 | 7888475 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os02g14370 |
| SBS | Chr3 | 21426818 | C-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 32111876 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 7242617 | T-A | HOMO | INTERGENIC | |
| SBS | Chr5 | 21310048 | T-C | HET | INTERGENIC | |
| SBS | Chr5 | 2165124 | A-G | HOMO | UTR_5_PRIME | |
| SBS | Chr5 | 5443790 | A-T | HOMO | INTRON | |
| SBS | Chr6 | 17681507 | A-T | HET | STOP_GAINED | LOC_Os06g30560 |
| SBS | Chr6 | 28975124 | T-C | HET | INTERGENIC | |
| SBS | Chr7 | 18054476 | T-C | HET | INTERGENIC | |
| SBS | Chr8 | 16935086 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os08g27800 |
| SBS | Chr9 | 18873283 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os09g31390 |
Deletions: 24
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr6 | 2360186 | 2360194 | 8 | LOC_Os06g05240 |
| Deletion | Chr2 | 2405868 | 2405877 | 9 | |
| Deletion | Chr11 | 6871981 | 6871982 | 1 | |
| Deletion | Chr1 | 7980001 | 8177000 | 197000 | 34 |
| Deletion | Chr11 | 8119974 | 8121702 | 1728 | LOC_Os11g14460 |
| Deletion | Chr3 | 9220134 | 9220144 | 10 | |
| Deletion | Chr10 | 9284922 | 9284933 | 11 | |
| Deletion | Chr4 | 9428313 | 9428338 | 25 | |
| Deletion | Chr3 | 12094166 | 12094167 | 1 | |
| Deletion | Chr3 | 15144476 | 15144477 | 1 | |
| Deletion | Chr3 | 16653261 | 16653262 | 1 | |
| Deletion | Chr1 | 16917487 | 16917492 | 5 | |
| Deletion | Chr5 | 18483798 | 18483799 | 1 | |
| Deletion | Chr7 | 22570266 | 22570272 | 6 | |
| Deletion | Chr3 | 26515712 | 26515714 | 2 | |
| Deletion | Chr5 | 26545623 | 26545625 | 2 | |
| Deletion | Chr8 | 26892459 | 26892497 | 38 | LOC_Os08g42550 |
| Deletion | Chr7 | 28566829 | 28568369 | 1540 | LOC_Os07g47840 |
| Deletion | Chr1 | 28777133 | 28777150 | 17 | |
| Deletion | Chr2 | 29173512 | 29173513 | 1 | LOC_Os02g47720 |
| Deletion | Chr5 | 29395001 | 29487000 | 92000 | 16 |
| Deletion | Chr4 | 30244377 | 30251535 | 7158 | 2 |
| Deletion | Chr3 | 32733895 | 32733919 | 24 | |
| Deletion | Chr3 | 35358322 | 35358325 | 3 |
Insertions: 3
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 31696031 | 31696031 | 1 | |
| Insertion | Chr2 | 590485 | 590485 | 1 | |
| Insertion | Chr6 | 20888479 | 20888480 | 2 |
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr3 | 12208057 | 13129719 | LOC_Os03g22730 |
| Inversion | Chr11 | 28760023 | 28873909 |
No Translocation
