Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN-137 [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN-137 Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 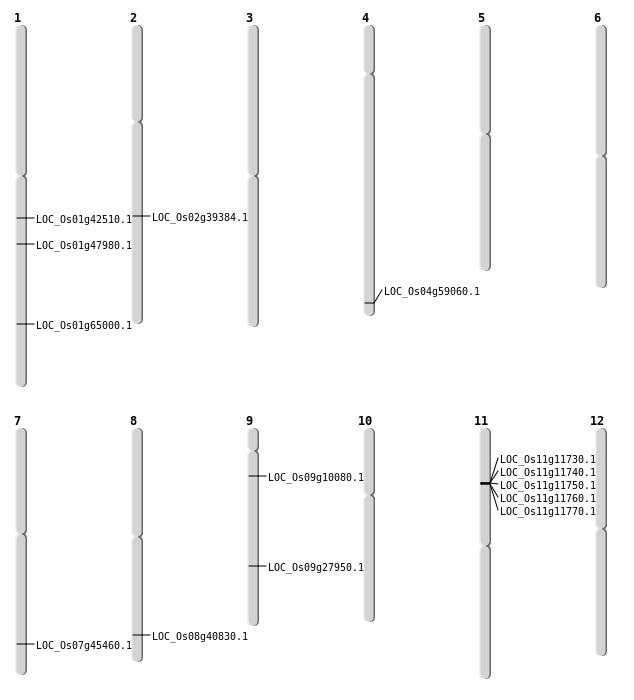
|
| Variant Information |
|---|
Single base substitutions: 33
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 13655777 | G-C | HOMO | INTERGENIC | |
| SBS | Chr1 | 17833819 | G-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 20335370 | C-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 37735395 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os01g65000 |
| SBS | Chr1 | 40421484 | C-A | HET | INTERGENIC | |
| SBS | Chr12 | 10271759 | G-T | HOMO | INTERGENIC | |
| SBS | Chr12 | 12642721 | T-G | HOMO | INTRON | |
| SBS | Chr12 | 26482482 | A-G | HOMO | INTRON | |
| SBS | Chr2 | 12169949 | T-G | HOMO | INTRON | |
| SBS | Chr2 | 34984688 | G-A | HET | INTRON | |
| SBS | Chr2 | 34984689 | T-A | HET | INTRON | |
| SBS | Chr3 | 14376926 | A-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 20975446 | C-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 35249024 | A-T | HET | INTERGENIC | |
| SBS | Chr3 | 35249025 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 19478056 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 23350689 | G-C | HET | INTERGENIC | |
| SBS | Chr4 | 28695580 | C-T | HOMO | INTERGENIC | |
| SBS | Chr4 | 35129238 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g59060 |
| SBS | Chr5 | 19428879 | A-G | HOMO | INTERGENIC | |
| SBS | Chr5 | 337222 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 16196322 | A-T | HOMO | INTERGENIC | |
| SBS | Chr6 | 185206 | A-G | HET | INTERGENIC | |
| SBS | Chr7 | 17032419 | G-A | HET | INTRON | |
| SBS | Chr7 | 7102858 | A-T | HET | INTERGENIC | |
| SBS | Chr7 | 9360913 | G-A | HET | INTRON | |
| SBS | Chr8 | 12324413 | C-T | HOMO | INTERGENIC | |
| SBS | Chr8 | 21272546 | T-C | HET | INTERGENIC | |
| SBS | Chr8 | 25838273 | T-G | HET | NON_SYNONYMOUS_CODING | LOC_Os08g40830 |
| SBS | Chr9 | 11930005 | T-G | HET | INTERGENIC | |
| SBS | Chr9 | 14529718 | T-A | HET | INTERGENIC | |
| SBS | Chr9 | 21285010 | G-T | HET | INTERGENIC | |
| SBS | Chr9 | 5495450 | G-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os09g10080 |
Deletions: 20
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr1 | 20624 | 20625 | 1 | |
| Deletion | Chr1 | 522132 | 522138 | 6 | |
| Deletion | Chr5 | 1453118 | 1453125 | 7 | |
| Deletion | Chr8 | 5190540 | 5190547 | 7 | |
| Deletion | Chr7 | 10561370 | 10561379 | 9 | |
| Deletion | Chr1 | 11708427 | 11708431 | 4 | |
| Deletion | Chr8 | 12903377 | 12903382 | 5 | |
| Deletion | Chr1 | 13370578 | 13370580 | 2 | |
| Deletion | Chr11 | 15708487 | 15708488 | 1 | |
| Deletion | Chr5 | 16604576 | 16604583 | 7 | |
| Deletion | Chr9 | 16983783 | 16983784 | 1 | LOC_Os09g27950 |
| Deletion | Chr8 | 17615128 | 17615138 | 10 | |
| Deletion | Chr3 | 18012357 | 18012358 | 1 | |
| Deletion | Chr4 | 19571799 | 19571800 | 1 | |
| Deletion | Chr3 | 23160200 | 23160201 | 1 | |
| Deletion | Chr6 | 23274534 | 23274541 | 7 | |
| Deletion | Chr1 | 24169805 | 24169806 | 1 | LOC_Os01g42510 |
| Deletion | Chr12 | 26482485 | 26482491 | 6 | |
| Deletion | Chr7 | 27123312 | 27123314 | 2 | LOC_Os07g45460 |
| Deletion | Chr2 | 32619596 | 32619601 | 5 |
Insertions: 3
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr12 | 23490639 | 23490640 | 2 | |
| Insertion | Chr6 | 12859888 | 12859888 | 1 | |
| Insertion | Chr6 | 3016076 | 3016076 | 1 |
No Inversion
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr2 | 23768571 | Chr1 | 27457623 | 2 |
| Translocation | Chr2 | 23768873 | Chr1 | 27457631 | 2 |
Tandem duplications: 1
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Tandem Duplication | Chr11 | 6519809 | 6538181 | 18372 | 5 |
