Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN-142 [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN-142 Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 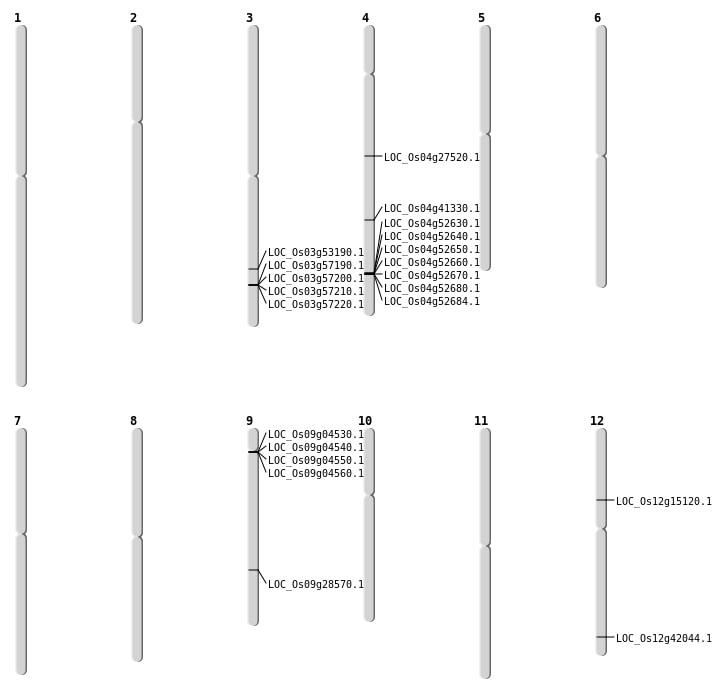
|
| Variant Information |
|---|
Single base substitutions: 34
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 11929906 | G-C | HET | INTERGENIC | |
| SBS | Chr1 | 2223140 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 22708561 | G-A | HOMO | INTERGENIC | |
| SBS | Chr1 | 28681953 | C-T | HOMO | INTERGENIC | |
| SBS | Chr10 | 3809088 | T-A | HET | INTERGENIC | |
| SBS | Chr11 | 10265783 | C-G | HET | INTRON | |
| SBS | Chr11 | 8291868 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 14676904 | C-A | HET | INTERGENIC | |
| SBS | Chr12 | 16130772 | C-G | HET | INTRON | |
| SBS | Chr12 | 25558978 | G-A | HET | UTR_3_PRIME | |
| SBS | Chr12 | 8626714 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g15120 |
| SBS | Chr2 | 17501427 | G-A | HET | INTRON | |
| SBS | Chr2 | 27076671 | A-G | HET | INTERGENIC | |
| SBS | Chr3 | 10228821 | C-A | HET | INTERGENIC | |
| SBS | Chr3 | 30163860 | G-A | HET | INTRON | |
| SBS | Chr3 | 34996627 | A-T | HET | UTR_5_PRIME | |
| SBS | Chr3 | 34996628 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 16110373 | C-A | HET | INTERGENIC | |
| SBS | Chr4 | 16269188 | G-C | HOMO | NON_SYNONYMOUS_CODING | LOC_Os04g27520 |
| SBS | Chr4 | 18724775 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 24017693 | T-A | HET | INTERGENIC | |
| SBS | Chr4 | 24526899 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g41330 |
| SBS | Chr4 | 545004 | A-T | HOMO | INTERGENIC | |
| SBS | Chr5 | 2018313 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 22032061 | A-G | HOMO | INTERGENIC | |
| SBS | Chr6 | 14616789 | C-G | HOMO | UTR_5_PRIME | |
| SBS | Chr6 | 15038282 | C-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 29639996 | A-G | HET | INTERGENIC | |
| SBS | Chr7 | 6960099 | A-G | HET | INTERGENIC | |
| SBS | Chr9 | 17385020 | T-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr9 | 17385025 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os09g28570 |
| SBS | Chr9 | 19188999 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 6049250 | T-C | HOMO | UTR_5_PRIME | |
| SBS | Chr9 | 6531282 | C-T | HET | INTERGENIC |
Deletions: 20
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr5 | 492225 | 492231 | 6 | |
| Deletion | Chr8 | 1814024 | 1814038 | 14 | |
| Deletion | Chr9 | 2419001 | 2435000 | 16000 | 4 |
| Deletion | Chr8 | 3239433 | 3239435 | 2 | |
| Deletion | Chr5 | 3365510 | 3365512 | 2 | |
| Deletion | Chr6 | 7461802 | 7462591 | 789 | |
| Deletion | Chr6 | 10319605 | 10319615 | 10 | |
| Deletion | Chr10 | 12665841 | 12665842 | 1 | |
| Deletion | Chr7 | 13144433 | 13144444 | 11 | |
| Deletion | Chr3 | 13931893 | 13931905 | 12 | |
| Deletion | Chr4 | 17891709 | 17891712 | 3 | |
| Deletion | Chr4 | 20349025 | 20349043 | 18 | |
| Deletion | Chr6 | 23321212 | 23321225 | 13 | |
| Deletion | Chr12 | 26084484 | 26084485 | 1 | LOC_Os12g42044 |
| Deletion | Chr2 | 26167685 | 26167713 | 28 | |
| Deletion | Chr5 | 28926059 | 28926061 | 2 | |
| Deletion | Chr4 | 31309001 | 31361000 | 52000 | 7 |
| Deletion | Chr4 | 31356027 | 31356438 | 411 | |
| Deletion | Chr3 | 32610001 | 32632000 | 22000 | 4 |
| Deletion | Chr1 | 42100401 | 42100405 | 4 |
Insertions: 3
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr10 | 12481382 | 12481382 | 1 | |
| Insertion | Chr12 | 25900257 | 25900258 | 2 | |
| Insertion | Chr4 | 7471318 | 7471318 | 1 |
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr4 | 31309729 | 31403326 | LOC_Os04g52630 |
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr4 | 31355992 | Chr3 | 30514572 | LOC_Os03g53190 |
| Translocation | Chr4 | 31403329 | Chr3 | 30513438 |
