Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN-143 [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN-143 Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 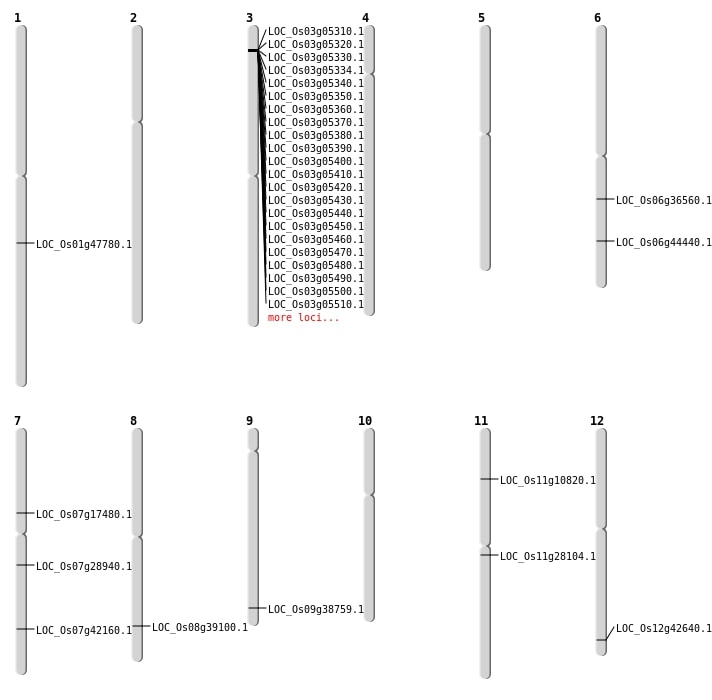
|
| Variant Information |
|---|
Single base substitutions: 21
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 12081194 | G-T | HET | INTERGENIC | |
| SBS | Chr1 | 35102780 | G-C | HET | INTERGENIC | |
| SBS | Chr1 | 8139761 | G-T | HOMO | INTERGENIC | |
| SBS | Chr10 | 1111079 | G-T | HET | INTERGENIC | |
| SBS | Chr11 | 28955954 | T-G | HOMO | INTRON | |
| SBS | Chr12 | 12889802 | G-A | HOMO | INTERGENIC | |
| SBS | Chr12 | 24952481 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 26503795 | T-G | HET | NON_SYNONYMOUS_CODING | LOC_Os12g42640 |
| SBS | Chr2 | 26492785 | T-A | HET | INTERGENIC | |
| SBS | Chr3 | 3013509 | A-T | HET | INTERGENIC | |
| SBS | Chr3 | 31358220 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 9004706 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os03g16310 |
| SBS | Chr6 | 13492022 | T-A | HET | INTERGENIC | |
| SBS | Chr6 | 18014600 | C-T | HOMO | INTERGENIC | |
| SBS | Chr6 | 26838388 | C-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os06g44440 |
| SBS | Chr7 | 10322632 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os07g17480 |
| SBS | Chr7 | 17258533 | T-C | HET | INTRON | |
| SBS | Chr7 | 25229062 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os07g42160 |
| SBS | Chr8 | 22701182 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 12677474 | C-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr9 | 18573040 | G-C | HET | INTRON |
Deletions: 15
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr3 | 2578001 | 2801000 | 223000 | 30 |
| Deletion | Chr6 | 9087004 | 9087015 | 11 | |
| Deletion | Chr7 | 16856597 | 16856606 | 9 | |
| Deletion | Chr2 | 16917147 | 16917160 | 13 | |
| Deletion | Chr2 | 18753679 | 18753683 | 4 | |
| Deletion | Chr6 | 21482728 | 21482729 | 1 | LOC_Os06g36560 |
| Deletion | Chr9 | 22273694 | 22273698 | 4 | LOC_Os09g38759 |
| Deletion | Chr8 | 24700706 | 24700707 | 1 | LOC_Os08g39100 |
| Deletion | Chr2 | 24927698 | 24927701 | 3 | |
| Deletion | Chr5 | 25797323 | 25797329 | 6 | |
| Deletion | Chr5 | 26094275 | 26094287 | 12 | |
| Deletion | Chr12 | 26560697 | 26560698 | 1 | |
| Deletion | Chr12 | 26718241 | 26718257 | 16 | |
| Deletion | Chr1 | 27341759 | 27341760 | 1 | LOC_Os01g47780 |
| Deletion | Chr1 | 35102659 | 35102676 | 17 |
Insertions: 3
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr2 | 24964446 | 24964446 | 1 | |
| Insertion | Chr7 | 23991181 | 23991181 | 1 | |
| Insertion | Chr9 | 13061771 | 13061772 | 2 |
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr11 | 5969345 | 6006801 | LOC_Os11g10820 |
Translocations: 3
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 5969104 | Chr7 | 16976311 | 2 |
| Translocation | Chr12 | 19511156 | Chr11 | 16160264 | LOC_Os11g28104 |
| Translocation | Chr12 | 19511265 | Chr11 | 16161621 |
