Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN-149 [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN-149 Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 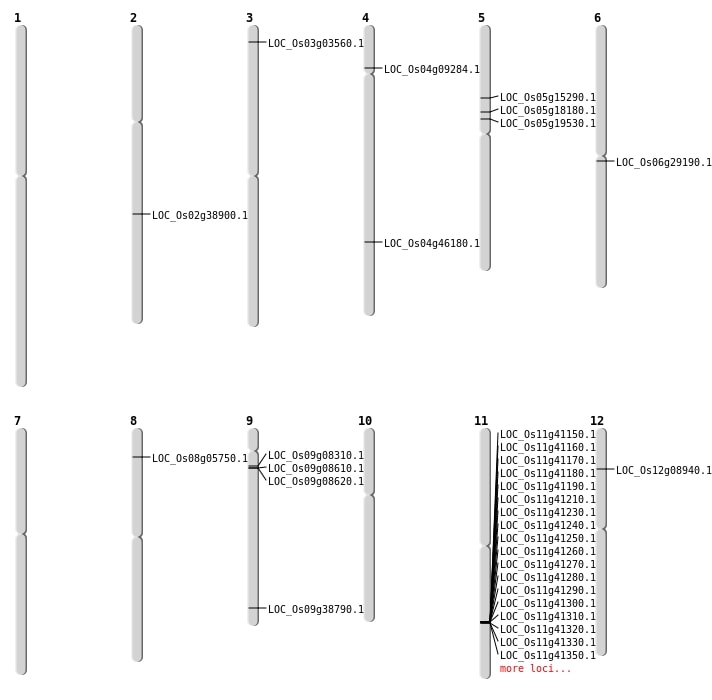
|
| Variant Information |
|---|
Single base substitutions: 29
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 2107632 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr1 | 29749815 | A-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 3887778 | T-C | HET | INTRON | |
| SBS | Chr1 | 5686503 | G-T | HET | INTERGENIC | |
| SBS | Chr1 | 7722852 | A-G | HET | INTERGENIC | |
| SBS | Chr10 | 14089602 | A-G | HET | INTERGENIC | |
| SBS | Chr10 | 15101096 | C-G | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr11 | 15361561 | C-G | HET | INTERGENIC | |
| SBS | Chr11 | 20050670 | G-A | HOMO | INTERGENIC | |
| SBS | Chr11 | 7785196 | G-A | HOMO | INTERGENIC | |
| SBS | Chr11 | 7785197 | A-G | HOMO | INTERGENIC | |
| SBS | Chr11 | 9394917 | G-A | HOMO | INTERGENIC | |
| SBS | Chr12 | 27524481 | A-T | HOMO | INTERGENIC | |
| SBS | Chr12 | 4668678 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g08940 |
| SBS | Chr3 | 15695146 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 19826674 | T-A | HET | INTERGENIC | |
| SBS | Chr3 | 25474782 | T-C | HET | INTERGENIC | |
| SBS | Chr4 | 13940734 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 19663786 | G-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 27355349 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g46180 |
| SBS | Chr5 | 16629385 | A-T | HOMO | INTERGENIC | |
| SBS | Chr5 | 8670060 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os05g15290 |
| SBS | Chr6 | 12556918 | T-C | HET | INTERGENIC | |
| SBS | Chr6 | 16658555 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os06g29190 |
| SBS | Chr6 | 16917794 | T-A | HET | INTERGENIC | |
| SBS | Chr7 | 4280835 | A-G | HET | INTERGENIC | |
| SBS | Chr8 | 22309031 | T-A | HET | INTERGENIC | |
| SBS | Chr8 | 5194750 | G-A | HOMO | INTERGENIC | |
| SBS | Chr9 | 5271313 | C-T | HET | INTERGENIC |
Deletions: 16
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr9 | 4485001 | 4508000 | 23000 | 2 |
| Deletion | Chr7 | 6661349 | 6661358 | 9 | |
| Deletion | Chr10 | 12185639 | 12185640 | 1 | |
| Deletion | Chr9 | 16715269 | 16715276 | 7 | |
| Deletion | Chr4 | 17702742 | 17702747 | 5 | |
| Deletion | Chr10 | 19073883 | 19073897 | 14 | |
| Deletion | Chr9 | 21168978 | 21168979 | 1 | |
| Deletion | Chr9 | 22287334 | 22287351 | 17 | LOC_Os09g38790 |
| Deletion | Chr2 | 23125595 | 23125599 | 4 | |
| Deletion | Chr2 | 23515528 | 23515533 | 5 | LOC_Os02g38900 |
| Deletion | Chr11 | 24652001 | 25051000 | 399000 | 54 |
| Deletion | Chr11 | 25075001 | 25656000 | 581000 | 82 |
| Deletion | Chr4 | 27355427 | 27355434 | 7 | LOC_Os04g46180 |
| Deletion | Chr4 | 29049873 | 29049879 | 6 | |
| Deletion | Chr2 | 32372040 | 32372041 | 1 | |
| Deletion | Chr4 | 32681717 | 32681736 | 19 |
Insertions: 2
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr10 | 2585702 | 2585704 | 3 | |
| Insertion | Chr5 | 8610648 | 8610650 | 3 |
Inversions: 3
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr8 | 3082045 | 3630910 | LOC_Os08g05750 |
| Inversion | Chr9 | 4297900 | 4508492 | LOC_Os09g08310 |
| Inversion | Chr5 | 10451957 | 11349782 | LOC_Os05g19530 |
Translocations: 4
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr8 | 3595884 | Chr3 | 1556650 | LOC_Os03g03560 |
| Translocation | Chr8 | 3630897 | Chr3 | 1556664 | LOC_Os03g03560 |
| Translocation | Chr5 | 10448337 | Chr4 | 5058852 | LOC_Os05g18180 |
| Translocation | Chr5 | 11349776 | Chr4 | 4933202 | 2 |
