Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN-164 [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN-164 Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 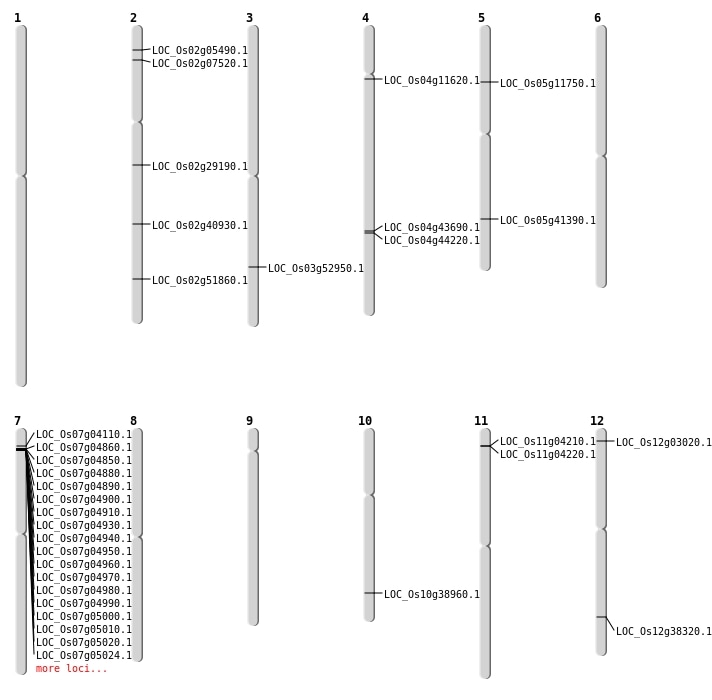
|
| Variant Information |
|---|
Single base substitutions: 37
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 15236785 | A-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 36117482 | G-A | HOMO | INTRON | |
| SBS | Chr10 | 15468601 | C-G | HOMO | INTERGENIC | |
| SBS | Chr10 | 1688173 | G-C | HOMO | INTERGENIC | |
| SBS | Chr10 | 20765509 | C-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os10g38960 |
| SBS | Chr10 | 21226608 | G-T | HOMO | UTR_3_PRIME | |
| SBS | Chr11 | 1172428 | A-G | HET | INTERGENIC | |
| SBS | Chr12 | 1129560 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os12g03020 |
| SBS | Chr12 | 8292296 | A-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 13843273 | C-T | HET | INTRON | |
| SBS | Chr2 | 3889748 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os02g07520 |
| SBS | Chr3 | 29722617 | G-T | HET | INTERGENIC | |
| SBS | Chr3 | 30372147 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os03g52950 |
| SBS | Chr3 | 4269098 | C-T | HET | INTRON | |
| SBS | Chr3 | 8147929 | C-G | HET | INTERGENIC | |
| SBS | Chr4 | 23624265 | G-T | HOMO | INTERGENIC | |
| SBS | Chr4 | 26181650 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g44220 |
| SBS | Chr4 | 34426439 | G-T | HOMO | INTERGENIC | |
| SBS | Chr4 | 6356967 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g11620 |
| SBS | Chr5 | 12101969 | A-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr5 | 15436276 | G-C | HET | INTERGENIC | |
| SBS | Chr5 | 24242839 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os05g41390 |
| SBS | Chr6 | 19583128 | A-T | HET | INTRON | |
| SBS | Chr6 | 20146627 | G-T | HET | INTRON | |
| SBS | Chr6 | 30968885 | T-C | HET | INTRON | |
| SBS | Chr7 | 13809645 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 15385210 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 17403130 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 19044128 | T-C | HOMO | INTERGENIC | |
| SBS | Chr7 | 19156138 | C-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 23939295 | T-A | HET | INTERGENIC | |
| SBS | Chr7 | 28181429 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os07g47120 |
| SBS | Chr7 | 5637664 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 6192930 | G-C | HET | INTRON | |
| SBS | Chr8 | 17030351 | G-A | HET | INTERGENIC | |
| SBS | Chr8 | 23625087 | T-C | HOMO | UTR_3_PRIME | |
| SBS | Chr9 | 1721287 | C-T | HET | INTERGENIC |
Deletions: 29
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr3 | 796958 | 796970 | 12 | |
| Deletion | Chr7 | 2144001 | 2390000 | 246000 | 42 |
| Deletion | Chr2 | 2656143 | 2656152 | 9 | LOC_Os02g05490 |
| Deletion | Chr1 | 2892440 | 2892441 | 1 | |
| Deletion | Chr10 | 2912160 | 2912176 | 16 | |
| Deletion | Chr6 | 4186862 | 4186872 | 10 | |
| Deletion | Chr9 | 4349164 | 4349165 | 1 | |
| Deletion | Chr5 | 6666606 | 6666610 | 4 | LOC_Os05g11750 |
| Deletion | Chr4 | 6911165 | 6911191 | 26 | |
| Deletion | Chr1 | 7658718 | 7658724 | 6 | |
| Deletion | Chr1 | 14140690 | 14140709 | 19 | |
| Deletion | Chr10 | 14179246 | 14179255 | 9 | |
| Deletion | Chr2 | 14609727 | 14609746 | 19 | |
| Deletion | Chr1 | 15239490 | 15239495 | 5 | |
| Deletion | Chr7 | 16506042 | 16506076 | 34 | |
| Deletion | Chr11 | 17738169 | 17738171 | 2 | |
| Deletion | Chr5 | 18706226 | 18706251 | 25 | |
| Deletion | Chr9 | 20892998 | 20893240 | 242 | |
| Deletion | Chr9 | 20893688 | 20893690 | 2 | |
| Deletion | Chr1 | 20907024 | 20907028 | 4 | |
| Deletion | Chr9 | 21177862 | 21180896 | 3034 | |
| Deletion | Chr12 | 24354103 | 24354152 | 49 | |
| Deletion | Chr4 | 25857839 | 25857863 | 24 | LOC_Os04g43690 |
| Deletion | Chr4 | 26468964 | 26468968 | 4 | |
| Deletion | Chr4 | 26620866 | 26620893 | 27 | |
| Deletion | Chr1 | 29195879 | 29195881 | 2 | |
| Deletion | Chr2 | 31768291 | 31768292 | 1 | LOC_Os02g51860 |
| Deletion | Chr3 | 34699945 | 34699949 | 4 | |
| Deletion | Chr3 | 35198477 | 35198487 | 10 |
Insertions: 3
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr11 | 28868362 | 28868363 | 2 | |
| Insertion | Chr11 | 6203339 | 6203342 | 4 | |
| Insertion | Chr12 | 23521192 | 23521195 | 4 | LOC_Os12g38320 |
Inversions: 5
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr11 | 219648 | 1724782 | LOC_Os11g04220 |
| Inversion | Chr7 | 1745641 | 2143339 | 2 |
| Inversion | Chr7 | 1745710 | 2389963 | 2 |
| Inversion | Chr2 | 16557809 | 17315895 | LOC_Os02g29190 |
| Inversion | Chr4 | 26090442 | 26242175 |
Translocations: 5
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr6 | 1124937 | Chr2 | 24803723 | LOC_Os02g40930 |
| Translocation | Chr6 | 1318987 | Chr2 | 24803946 | LOC_Os02g40930 |
| Translocation | Chr11 | 1717708 | Chr5 | 24088941 | LOC_Os11g04210 |
| Translocation | Chr10 | 22050947 | Chr9 | 20893227 | |
| Translocation | Chr10 | 22050951 | Chr9 | 20893004 |
