Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN-167 [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN-167 Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 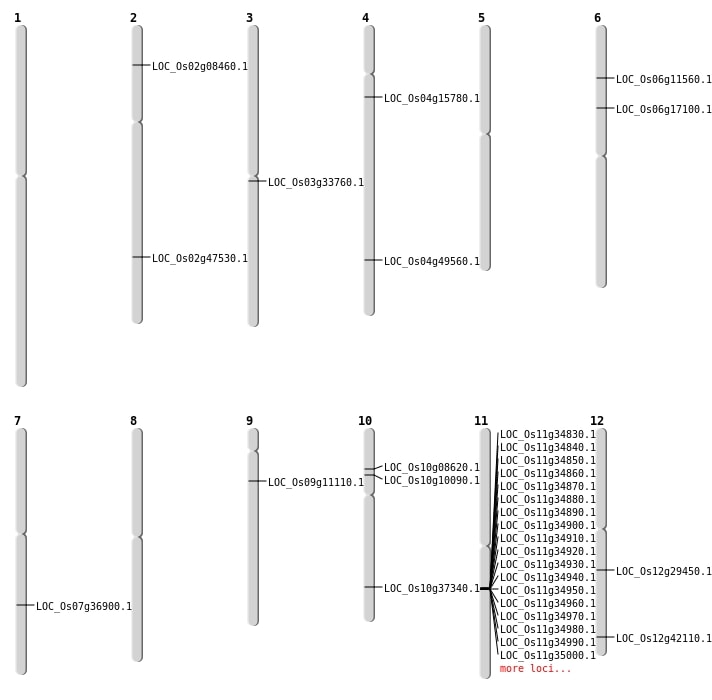
|
| Variant Information |
|---|
Single base substitutions: 35
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 28566540 | A-T | HOMO | INTERGENIC | |
| SBS | Chr10 | 4440214 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 1762454 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 16572466 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr12 | 19509480 | G-T | HOMO | INTRON | |
| SBS | Chr12 | 27369720 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 7240864 | A-C | HET | INTERGENIC | |
| SBS | Chr12 | 8954890 | G-T | HOMO | INTRON | |
| SBS | Chr2 | 11576598 | C-G | HET | INTERGENIC | |
| SBS | Chr2 | 14798106 | C-A | HET | INTERGENIC | |
| SBS | Chr2 | 34021908 | A-T | HET | INTERGENIC | |
| SBS | Chr2 | 5949418 | C-A | HET | INTERGENIC | |
| SBS | Chr2 | 5949419 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 15654858 | G-A | HET | INTRON | |
| SBS | Chr3 | 17963854 | A-G | HET | INTERGENIC | |
| SBS | Chr3 | 4141180 | T-A | HET | INTRON | |
| SBS | Chr3 | 4141181 | T-A | HET | INTRON | |
| SBS | Chr3 | 4141293 | A-G | HET | INTRON | |
| SBS | Chr4 | 10792584 | C-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr4 | 1538725 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 15863762 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 29557157 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os04g49560 |
| SBS | Chr4 | 5189255 | T-C | HET | INTERGENIC | |
| SBS | Chr4 | 5497576 | A-C | HET | INTERGENIC | |
| SBS | Chr6 | 5421478 | T-C | HET | INTRON | |
| SBS | Chr6 | 9989499 | A-T | HET | INTERGENIC | |
| SBS | Chr6 | 9989500 | A-T | HET | INTERGENIC | |
| SBS | Chr7 | 13867791 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 14026266 | C-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 22097045 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os07g36900 |
| SBS | Chr7 | 4130929 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 16603022 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr9 | 14346924 | G-C | HET | INTERGENIC | |
| SBS | Chr9 | 3981344 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 6162278 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os09g11110 |
Deletions: 29
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr4 | 609741 | 609742 | 1 | |
| Deletion | Chr10 | 635759 | 635763 | 4 | |
| Deletion | Chr4 | 3144577 | 3144584 | 7 | |
| Deletion | Chr2 | 4204270 | 4204276 | 6 | |
| Deletion | Chr2 | 4556631 | 4557827 | 1196 | LOC_Os02g08460 |
| Deletion | Chr3 | 5165120 | 5165186 | 66 | |
| Deletion | Chr4 | 5187320 | 5187321 | 1 | |
| Deletion | Chr5 | 5281316 | 5281321 | 5 | |
| Deletion | Chr6 | 6129209 | 6129216 | 7 | LOC_Os06g11560 |
| Deletion | Chr6 | 9912599 | 9914812 | 2213 | LOC_Os06g17100 |
| Deletion | Chr12 | 11754006 | 11754016 | 10 | |
| Deletion | Chr11 | 13225627 | 13225631 | 4 | |
| Deletion | Chr6 | 14266529 | 14266530 | 1 | |
| Deletion | Chr2 | 15760571 | 15760579 | 8 | |
| Deletion | Chr3 | 16204375 | 16204446 | 71 | |
| Deletion | Chr12 | 17497167 | 17497171 | 4 | LOC_Os12g29450 |
| Deletion | Chr5 | 18249535 | 18249548 | 13 | |
| Deletion | Chr3 | 19302504 | 19302514 | 10 | LOC_Os03g33760 |
| Deletion | Chr10 | 20002094 | 20011335 | 9241 | LOC_Os10g37340 |
| Deletion | Chr11 | 20410001 | 20626000 | 216000 | 32 |
| Deletion | Chr4 | 20574010 | 20574011 | 1 | |
| Deletion | Chr11 | 20633001 | 20663000 | 30000 | 6 |
| Deletion | Chr11 | 20667001 | 20713000 | 46000 | 6 |
| Deletion | Chr4 | 23632277 | 23632281 | 4 | |
| Deletion | Chr7 | 23744078 | 23744083 | 5 | |
| Deletion | Chr1 | 24120778 | 24120782 | 4 | |
| Deletion | Chr12 | 26801223 | 26801224 | 1 | |
| Deletion | Chr5 | 27402829 | 27402831 | 2 | |
| Deletion | Chr2 | 29039200 | 29039207 | 7 | LOC_Os02g47530 |
Insertions: 7
Inversions: 3
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr10 | 4613168 | 4682286 | LOC_Os10g08620 |
| Inversion | Chr10 | 4682291 | 5511266 | 2 |
| Inversion | Chr12 | 26115114 | 27279774 | LOC_Os12g42110 |
Translocations: 1
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr4 | 8573643 | Chr3 | 1730843 | LOC_Os04g15780 |
