Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN-170 [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN-170 Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 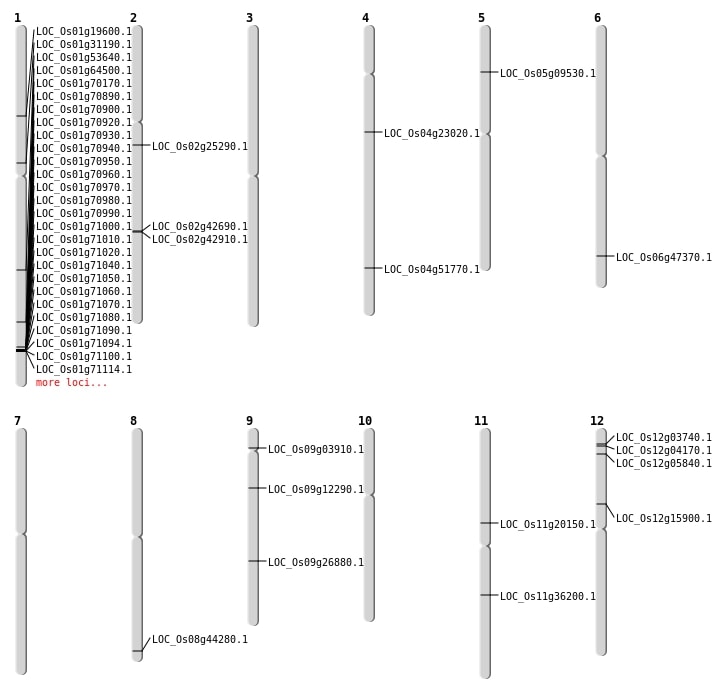
|
| Variant Information |
|---|
Single base substitutions: 49
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 11107716 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g19600 |
| SBS | Chr1 | 11927183 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 13528212 | T-G | HOMO | INTRON | |
| SBS | Chr1 | 17056545 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os01g31190 |
| SBS | Chr1 | 17267463 | A-T | HOMO | INTRON | |
| SBS | Chr1 | 17267471 | A-C | HOMO | INTRON | |
| SBS | Chr1 | 30821548 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os01g53640 |
| SBS | Chr1 | 30821549 | A-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr1 | 36032230 | A-C | HET | INTERGENIC | |
| SBS | Chr1 | 40611596 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os01g70170 |
| SBS | Chr1 | 40611600 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr1 | 8443596 | A-G | HET | INTERGENIC | |
| SBS | Chr1 | 8828234 | T-C | HET | INTERGENIC | |
| SBS | Chr10 | 18849951 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 18849953 | A-T | HET | INTERGENIC | |
| SBS | Chr11 | 11642319 | G-A | HET | STOP_GAINED | LOC_Os11g20150 |
| SBS | Chr11 | 12913892 | A-G | HET | INTERGENIC | |
| SBS | Chr11 | 8383437 | T-C | HET | INTERGENIC | |
| SBS | Chr12 | 14954085 | T-C | HOMO | INTERGENIC | |
| SBS | Chr12 | 2689143 | T-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os12g05840 |
| SBS | Chr12 | 2689144 | G-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr12 | 2689145 | A-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os12g05840 |
| SBS | Chr12 | 2689272 | A-C | HOMO | NON_SYNONYMOUS_CODING | LOC_Os12g05840 |
| SBS | Chr2 | 25674916 | A-C | HOMO | INTRON | |
| SBS | Chr2 | 25807047 | G-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os02g42910 |
| SBS | Chr3 | 13021165 | A-G | HET | INTERGENIC | |
| SBS | Chr3 | 14706544 | T-C | HET | INTERGENIC | |
| SBS | Chr3 | 23123639 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 30436313 | G-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 4430620 | A-G | HET | INTRON | |
| SBS | Chr4 | 16237743 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 25232268 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 25608256 | C-G | HET | UTR_3_PRIME | |
| SBS | Chr4 | 28657501 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 31926780 | C-G | HET | INTRON | |
| SBS | Chr4 | 33090473 | T-C | HET | INTERGENIC | |
| SBS | Chr5 | 13868244 | C-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr6 | 3993743 | T-C | HET | INTERGENIC | |
| SBS | Chr7 | 3626375 | A-G | HET | INTRON | |
| SBS | Chr7 | 4321447 | T-C | HET | INTERGENIC | |
| SBS | Chr8 | 12761266 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 21037451 | T-C | HET | INTERGENIC | |
| SBS | Chr8 | 22297841 | T-G | HET | INTRON | |
| SBS | Chr8 | 27878378 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os08g44280 |
| SBS | Chr8 | 4401552 | C-A | HET | INTERGENIC | |
| SBS | Chr9 | 13838883 | G-A | HOMO | INTRON | |
| SBS | Chr9 | 2000481 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os09g03910 |
| SBS | Chr9 | 20575411 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 21247932 | A-G | HET | INTERGENIC |
Deletions: 30
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr10 | 2156333 | 2156340 | 7 | |
| Deletion | Chr7 | 3626372 | 3626373 | 1 | |
| Deletion | Chr5 | 5390216 | 5390217 | 1 | LOC_Os05g09530 |
| Deletion | Chr5 | 5672254 | 5672282 | 28 | |
| Deletion | Chr6 | 5691817 | 5691826 | 9 | |
| Deletion | Chr5 | 7926794 | 7926798 | 4 | |
| Deletion | Chr4 | 8838395 | 8838396 | 1 | |
| Deletion | Chr11 | 9128712 | 9128718 | 6 | |
| Deletion | Chr3 | 10506315 | 10506316 | 1 | |
| Deletion | Chr11 | 13091414 | 13091415 | 1 | |
| Deletion | Chr7 | 13101569 | 13101588 | 19 | |
| Deletion | Chr2 | 14751123 | 14751124 | 1 | LOC_Os02g25290 |
| Deletion | Chr8 | 15193421 | 15193424 | 3 | |
| Deletion | Chr9 | 16338217 | 16338218 | 1 | LOC_Os09g26880 |
| Deletion | Chr9 | 16683497 | 16683510 | 13 | |
| Deletion | Chr4 | 18721975 | 18721992 | 17 | |
| Deletion | Chr1 | 19202259 | 19202260 | 1 | |
| Deletion | Chr1 | 19724419 | 19724438 | 19 | |
| Deletion | Chr11 | 21300032 | 21300033 | 1 | LOC_Os11g36200 |
| Deletion | Chr3 | 22872246 | 22872251 | 5 | |
| Deletion | Chr11 | 25099430 | 25099432 | 2 | |
| Deletion | Chr2 | 25674857 | 25674859 | 2 | LOC_Os02g42690 |
| Deletion | Chr7 | 25871490 | 25871493 | 3 | |
| Deletion | Chr12 | 26836625 | 26836635 | 10 | |
| Deletion | Chr5 | 27983062 | 27983068 | 6 | |
| Deletion | Chr6 | 28711656 | 28711658 | 2 | LOC_Os06g47370 |
| Deletion | Chr3 | 30186777 | 30186779 | 2 | |
| Deletion | Chr4 | 30678941 | 30678948 | 7 | LOC_Os04g51770 |
| Deletion | Chr1 | 36154388 | 36154392 | 4 | |
| Deletion | Chr1 | 41028284 | 41246188 | 217904 | 40 |
No Insertion
Inversions: 3
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr12 | 1021618 | 1751829 | LOC_Os12g04170 |
| Inversion | Chr12 | 1515711 | 1751827 | 2 |
| Inversion | Chr9 | 7003343 | 8344656 | LOC_Os09g12290 |
Translocations: 3
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 9052713 | Chr4 | 13069486 | 2 |
| Translocation | Chr12 | 15822761 | Chr1 | 37426820 | LOC_Os01g64500 |
| Translocation | Chr12 | 15823022 | Chr1 | 37426736 | LOC_Os01g64500 |
