Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN-175 [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN-175 Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 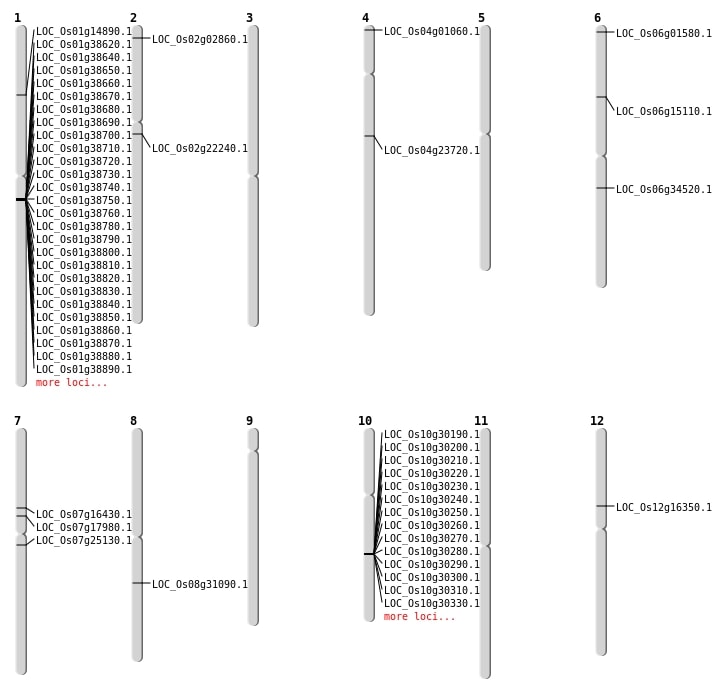
|
| Variant Information |
|---|
Single base substitutions: 47
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 10911213 | C-A | HET | INTERGENIC | |
| SBS | Chr1 | 10911214 | T-C | HET | INTERGENIC | |
| SBS | Chr1 | 10976996 | T-C | HET | INTERGENIC | |
| SBS | Chr1 | 10990125 | C-A | HET | INTERGENIC | |
| SBS | Chr1 | 11708662 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 17253620 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 19836834 | G-A | HET | INTRON | |
| SBS | Chr1 | 35333861 | T-C | HET | INTERGENIC | |
| SBS | Chr1 | 39296421 | T-A | HET | UTR_3_PRIME | |
| SBS | Chr1 | 8344476 | C-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os01g14890 |
| SBS | Chr10 | 3543713 | C-A | HET | INTERGENIC | |
| SBS | Chr10 | 4732797 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 8107673 | C-T | HOMO | INTERGENIC | |
| SBS | Chr11 | 14151197 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 21637471 | C-G | HOMO | INTERGENIC | |
| SBS | Chr11 | 231642 | A-G | HET | UTR_3_PRIME | |
| SBS | Chr12 | 5309297 | A-C | HET | INTERGENIC | |
| SBS | Chr12 | 9414277 | A-T | HET | INTERGENIC | |
| SBS | Chr2 | 1090872 | A-C | HET | NON_SYNONYMOUS_CODING | LOC_Os02g02860 |
| SBS | Chr2 | 1826165 | T-G | HET | UTR_3_PRIME | |
| SBS | Chr2 | 23166301 | C-G | HOMO | INTERGENIC | |
| SBS | Chr2 | 2634269 | T-A | HET | INTERGENIC | |
| SBS | Chr2 | 28428873 | T-C | HOMO | INTERGENIC | |
| SBS | Chr2 | 33444664 | T-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 33444665 | T-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 18647820 | G-T | HET | INTERGENIC | |
| SBS | Chr4 | 31289303 | T-C | HET | UTR_5_PRIME | |
| SBS | Chr4 | 33927516 | A-T | HET | INTERGENIC | |
| SBS | Chr4 | 4947892 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr5 | 11700425 | C-A | HET | INTRON | |
| SBS | Chr5 | 18810407 | A-C | HET | INTERGENIC | |
| SBS | Chr5 | 19351564 | G-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr5 | 20720234 | A-T | HOMO | INTERGENIC | |
| SBS | Chr5 | 24722730 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 9653319 | C-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr6 | 13315437 | C-A | HET | INTRON | |
| SBS | Chr6 | 8564446 | G-A | HOMO | STOP_GAINED | LOC_Os06g15110 |
| SBS | Chr7 | 10647379 | C-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os07g17980 |
| SBS | Chr7 | 14359989 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os07g25130 |
| SBS | Chr7 | 22348933 | T-A | HET | INTERGENIC | |
| SBS | Chr7 | 22596805 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 24056222 | T-G | HET | INTERGENIC | |
| SBS | Chr7 | 24602081 | A-G | HET | INTRON | |
| SBS | Chr7 | 24641655 | A-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 9624352 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os07g16430 |
| SBS | Chr8 | 18808807 | C-T | HOMO | INTRON | |
| SBS | Chr8 | 3272385 | G-A | HET | INTERGENIC |
Deletions: 29
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr4 | 53040 | 53042 | 2 | LOC_Os04g01060 |
| Deletion | Chr6 | 344277 | 344286 | 9 | LOC_Os06g01580 |
| Deletion | Chr4 | 1005633 | 1005635 | 2 | |
| Deletion | Chr4 | 1420648 | 1420776 | 128 | |
| Deletion | Chr6 | 2942664 | 2942679 | 15 | |
| Deletion | Chr12 | 4734415 | 4734416 | 1 | |
| Deletion | Chr12 | 6680711 | 6680713 | 2 | |
| Deletion | Chr12 | 7495524 | 7495526 | 2 | |
| Deletion | Chr4 | 7678552 | 7678556 | 4 | |
| Deletion | Chr7 | 7972897 | 7972898 | 1 | |
| Deletion | Chr12 | 9357007 | 9357009 | 2 | LOC_Os12g16350 |
| Deletion | Chr9 | 10187051 | 10187052 | 1 | |
| Deletion | Chr8 | 11919710 | 11919713 | 3 | |
| Deletion | Chr2 | 13262366 | 13262373 | 7 | LOC_Os02g22240 |
| Deletion | Chr4 | 13568850 | 13568852 | 2 | LOC_Os04g23720 |
| Deletion | Chr10 | 15689868 | 15939803 | 249935 | 40 |
| Deletion | Chr6 | 16013231 | 16013232 | 1 | |
| Deletion | Chr7 | 17906283 | 17906285 | 2 | |
| Deletion | Chr11 | 18748391 | 18748434 | 43 | |
| Deletion | Chr8 | 19206984 | 19206985 | 1 | LOC_Os08g31090 |
| Deletion | Chr9 | 19533727 | 19533732 | 5 | |
| Deletion | Chr1 | 21691488 | 21857241 | 165753 | 29 |
| Deletion | Chr1 | 21794546 | 21794547 | 1 | |
| Deletion | Chr1 | 21847305 | 21847308 | 3 | |
| Deletion | Chr4 | 23014572 | 23014574 | 2 | |
| Deletion | Chr3 | 24965270 | 24965272 | 2 | |
| Deletion | Chr1 | 28500774 | 28500775 | 1 | |
| Deletion | Chr4 | 33783153 | 33783205 | 52 | |
| Deletion | Chr1 | 35226988 | 35226999 | 11 |
Insertions: 2
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr10 | 18655784 | 18655785 | 2 | |
| Insertion | Chr4 | 26538353 | 26538353 | 1 |
No Inversion
Translocations: 3
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr10 | 19357944 | Chr6 | 20085140 | LOC_Os10g36210 |
| Translocation | Chr10 | 19357986 | Chr6 | 20086566 | 2 |
| Translocation | Chr11 | 23286499 | Chr1 | 41587095 |
