Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN-181 [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN-181 Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 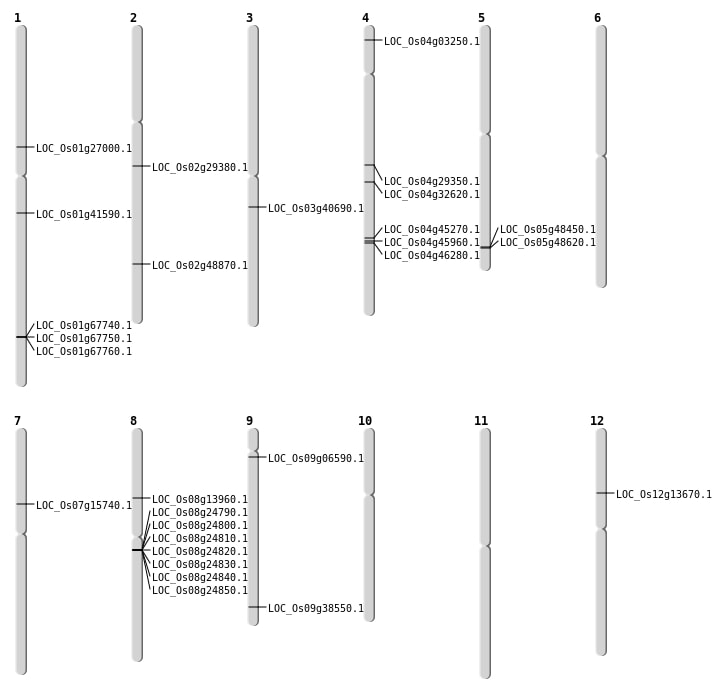
|
| Variant Information |
|---|
Single base substitutions: 45
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 1431755 | T-A | HET | INTERGENIC | |
| SBS | Chr1 | 14460269 | A-G | HET | SYNONYMOUS_CODING | |
| SBS | Chr1 | 14559156 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr1 | 15043653 | C-G | HOMO | NON_SYNONYMOUS_CODING | LOC_Os01g27000 |
| SBS | Chr1 | 3085177 | A-G | HET | INTERGENIC | |
| SBS | Chr1 | 33454440 | G-T | HOMO | INTRON | |
| SBS | Chr10 | 13331810 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 1627253 | A-G | HOMO | INTRON | |
| SBS | Chr11 | 27227555 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 11345655 | T-C | HET | INTERGENIC | |
| SBS | Chr12 | 4420677 | A-C | HOMO | INTERGENIC | |
| SBS | Chr12 | 7697343 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os12g13670 |
| SBS | Chr12 | 7955353 | T-C | HET | INTRON | |
| SBS | Chr2 | 12564491 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 12848142 | A-G | HET | INTRON | |
| SBS | Chr2 | 27054360 | A-C | HOMO | INTRON | |
| SBS | Chr2 | 33649187 | A-T | HET | INTERGENIC | |
| SBS | Chr2 | 33649188 | A-T | HET | INTERGENIC | |
| SBS | Chr2 | 4134492 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 7728039 | T-A | HET | INTERGENIC | |
| SBS | Chr3 | 20575485 | T-C | HET | INTRON | |
| SBS | Chr3 | 20661411 | C-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 22627252 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os03g40690 |
| SBS | Chr3 | 23251196 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 29137379 | C-A | HET | INTERGENIC | |
| SBS | Chr3 | 29395258 | G-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 35903866 | A-T | HOMO | INTERGENIC | |
| SBS | Chr4 | 1387689 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os04g03250 |
| SBS | Chr4 | 27648631 | G-C | HET | INTERGENIC | |
| SBS | Chr4 | 8384154 | G-A | HET | INTRON | |
| SBS | Chr5 | 10582488 | T-G | HET | INTERGENIC | |
| SBS | Chr5 | 18483109 | G-T | HET | INTERGENIC | |
| SBS | Chr5 | 23940920 | G-A | HET | INTRON | |
| SBS | Chr5 | 9580772 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 270727 | C-T | HET | INTRON | |
| SBS | Chr7 | 10475206 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 1233290 | T-C | HET | INTRON | |
| SBS | Chr7 | 18234829 | C-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 25680276 | A-C | HET | INTERGENIC | |
| SBS | Chr7 | 4750969 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr7 | 9140633 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os07g15740 |
| SBS | Chr8 | 3837447 | G-T | HET | INTERGENIC | |
| SBS | Chr9 | 3132341 | G-T | HET | STOP_GAINED | LOC_Os09g06590 |
| SBS | Chr9 | 5353288 | T-A | HET | INTERGENIC | |
| SBS | Chr9 | 6221996 | T-A | HET | SYNONYMOUS_CODING |
Deletions: 14
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr8 | 8370442 | 8370447 | 5 | LOC_Os08g13960 |
| Deletion | Chr8 | 15008001 | 15040000 | 32000 | 7 |
| Deletion | Chr4 | 16097357 | 16097362 | 5 | |
| Deletion | Chr3 | 19533816 | 19533817 | 1 | |
| Deletion | Chr8 | 19888969 | 19888970 | 1 | |
| Deletion | Chr5 | 20447119 | 20447120 | 1 | |
| Deletion | Chr9 | 22176589 | 22176596 | 7 | LOC_Os09g38550 |
| Deletion | Chr8 | 23269941 | 23269943 | 2 | |
| Deletion | Chr1 | 23548076 | 23548362 | 286 | LOC_Os01g41590 |
| Deletion | Chr11 | 24087075 | 24087104 | 29 | |
| Deletion | Chr7 | 26435511 | 26435520 | 9 | |
| Deletion | Chr4 | 26748000 | 26748003 | 3 | LOC_Os04g45270 |
| Deletion | Chr2 | 29894260 | 29894263 | 3 | LOC_Os02g48870 |
| Deletion | Chr1 | 39374001 | 39384000 | 10000 | 3 |
Insertions: 5
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 39916646 | 39916649 | 4 | |
| Insertion | Chr2 | 17454917 | 17454918 | 2 | LOC_Os02g29380 |
| Insertion | Chr3 | 2462821 | 2462821 | 1 | |
| Insertion | Chr4 | 27851674 | 27851674 | 1 | |
| Insertion | Chr5 | 17505271 | 17505272 | 2 |
Inversions: 4
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr4 | 17427221 | 17438097 | LOC_Os04g29350 |
| Inversion | Chr4 | 17438115 | 17445586 | |
| Inversion | Chr4 | 27222856 | 27425251 | 2 |
| Inversion | Chr5 | 27779856 | 27869684 | 2 |
Translocations: 1
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 7988832 | Chr4 | 19655126 | LOC_Os04g32620 |
Tandem duplications: 1
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Tandem Duplication | Chr3 | 36136483 | 36140436 | 3953 |
