Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN-183 [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN-183 Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 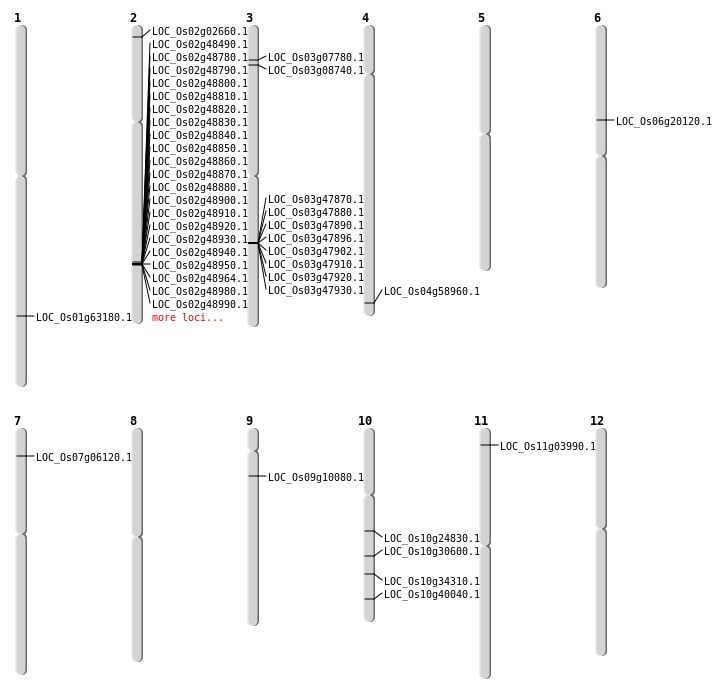
|
| Variant Information |
|---|
Single base substitutions: 28
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 31045966 | C-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 36620280 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g63180 |
| SBS | Chr10 | 12770940 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os10g24830 |
| SBS | Chr10 | 21444894 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os10g40040 |
| SBS | Chr12 | 10271759 | G-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 15090141 | A-G | HET | INTRON | |
| SBS | Chr2 | 15652405 | G-A | HET | INTRON | |
| SBS | Chr2 | 20137359 | C-T | HET | INTRON | |
| SBS | Chr2 | 29685399 | T-G | HET | INTRON | |
| SBS | Chr2 | 35132292 | A-T | HET | INTERGENIC | |
| SBS | Chr2 | 978353 | A-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os02g02660 |
| SBS | Chr2 | 978354 | G-T | HOMO | STOP_GAINED | LOC_Os02g02660 |
| SBS | Chr3 | 1941734 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 29287701 | A-T | HET | INTERGENIC | |
| SBS | Chr3 | 30906612 | A-G | HET | INTRON | |
| SBS | Chr4 | 15230289 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 22471492 | T-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 3709760 | C-T | HOMO | INTERGENIC | |
| SBS | Chr5 | 16671997 | A-T | HET | INTERGENIC | |
| SBS | Chr5 | 9380481 | G-A | HOMO | INTERGENIC | |
| SBS | Chr6 | 16884905 | T-A | HET | INTERGENIC | |
| SBS | Chr7 | 11953069 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr7 | 12454205 | T-C | HET | INTRON | |
| SBS | Chr7 | 24692276 | T-C | HET | INTERGENIC | |
| SBS | Chr7 | 6051024 | A-G | HET | INTERGENIC | |
| SBS | Chr8 | 9711615 | A-G | HET | INTERGENIC | |
| SBS | Chr9 | 1377531 | G-A | HOMO | INTERGENIC | |
| SBS | Chr9 | 5495450 | G-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os09g10080 |
Deletions: 23
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr11 | 1610991 | 1611006 | 15 | LOC_Os11g03990 |
| Deletion | Chr7 | 2969374 | 2969381 | 7 | LOC_Os07g06120 |
| Deletion | Chr1 | 4024798 | 4024810 | 12 | |
| Deletion | Chr6 | 6835625 | 6835626 | 1 | |
| Deletion | Chr9 | 7021966 | 7021993 | 27 | |
| Deletion | Chr6 | 7919775 | 7919799 | 24 | |
| Deletion | Chr10 | 8047226 | 8047227 | 1 | |
| Deletion | Chr7 | 9070906 | 9070907 | 1 | |
| Deletion | Chr6 | 11537403 | 11537404 | 1 | LOC_Os06g20120 |
| Deletion | Chr10 | 15921102 | 15921116 | 14 | LOC_Os10g30600 |
| Deletion | Chr1 | 17940936 | 17940937 | 1 | |
| Deletion | Chr10 | 18299352 | 18299355 | 3 | LOC_Os10g34310 |
| Deletion | Chr4 | 22111848 | 22111855 | 7 | |
| Deletion | Chr5 | 23670447 | 23670454 | 7 | |
| Deletion | Chr11 | 24070436 | 24070438 | 2 | |
| Deletion | Chr3 | 25379193 | 25379210 | 17 | |
| Deletion | Chr8 | 25431929 | 25431931 | 2 | |
| Deletion | Chr3 | 27213914 | 27239560 | 25646 | 8 |
| Deletion | Chr3 | 27550363 | 27550376 | 13 | |
| Deletion | Chr2 | 29854001 | 30122000 | 268000 | 49 |
| Deletion | Chr2 | 30109600 | 30138579 | 28979 | 6 |
| Deletion | Chr4 | 35074674 | 35074677 | 3 | LOC_Os04g58960 |
| Deletion | Chr1 | 39878436 | 39878438 | 2 |
Insertions: 2
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 22761873 | 22761875 | 3 | |
| Insertion | Chr11 | 12560722 | 12560722 | 1 |
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr3 | 3952285 | 4017068 | LOC_Os03g07780 |
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr3 | 3952289 | Chr2 | 29685375 | 2 |
| Translocation | Chr3 | 4517102 | Chr2 | 29854343 | 2 |
