Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN-189 [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN-189 Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 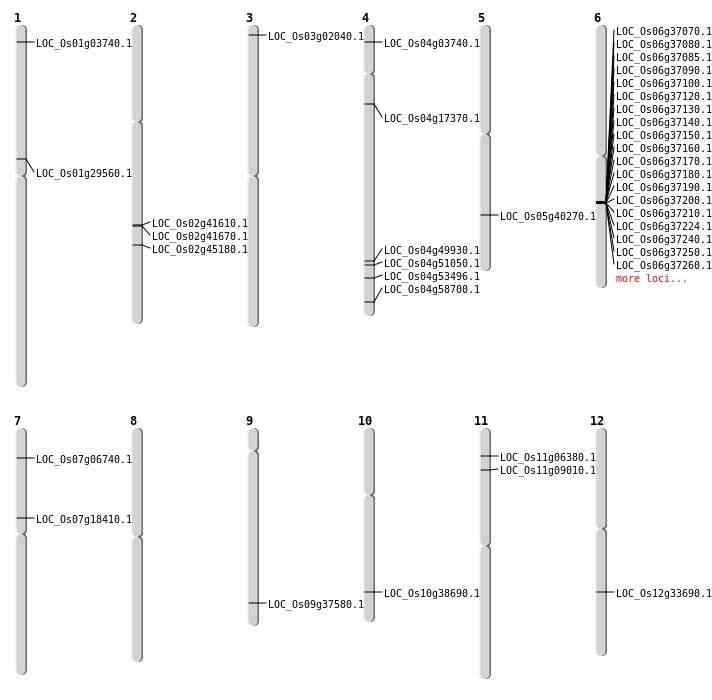
|
| Variant Information |
|---|
Single base substitutions: 48
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 10696927 | C-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 16569387 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os01g29560 |
| SBS | Chr1 | 31847859 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 34598488 | A-G | HET | INTRON | |
| SBS | Chr1 | 38111373 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 38111374 | A-T | HET | INTERGENIC | |
| SBS | Chr1 | 7264737 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 20541353 | T-A | HET | INTERGENIC | |
| SBS | Chr10 | 21165216 | T-A | HET | INTERGENIC | |
| SBS | Chr11 | 10294592 | C-G | HOMO | INTERGENIC | |
| SBS | Chr11 | 18323099 | C-A | HET | INTERGENIC | |
| SBS | Chr11 | 3067878 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os11g06380 |
| SBS | Chr11 | 6915677 | C-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr12 | 25929515 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 4942168 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 928467 | A-T | HET | INTERGENIC | |
| SBS | Chr2 | 27524755 | A-T | HET | INTERGENIC | |
| SBS | Chr3 | 25605571 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 28200544 | C-A | HET | INTERGENIC | |
| SBS | Chr3 | 9045918 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 1653038 | T-G | HET | NON_SYNONYMOUS_CODING | LOC_Os04g03740 |
| SBS | Chr4 | 17171017 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 19719919 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 21381342 | T-C | HOMO | INTRON | |
| SBS | Chr4 | 30221149 | T-G | HOMO | NON_SYNONYMOUS_CODING | LOC_Os04g51050 |
| SBS | Chr4 | 32063884 | G-A | HOMO | UTR_5_PRIME | |
| SBS | Chr4 | 34349722 | A-G | HET | INTRON | |
| SBS | Chr4 | 9513447 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g17370 |
| SBS | Chr5 | 1982016 | C-T | HOMO | INTERGENIC | |
| SBS | Chr5 | 23660510 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os05g40270 |
| SBS | Chr5 | 4794471 | C-T | HOMO | INTERGENIC | |
| SBS | Chr5 | 5244113 | C-T | HOMO | INTRON | |
| SBS | Chr6 | 24180715 | C-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr7 | 10900669 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os07g18410 |
| SBS | Chr7 | 12962864 | C-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 22710315 | G-A | HOMO | INTERGENIC | |
| SBS | Chr7 | 28169912 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 8914668 | G-T | HET | INTERGENIC | |
| SBS | Chr8 | 11186889 | A-G | HET | INTERGENIC | |
| SBS | Chr8 | 18120331 | T-C | HET | INTERGENIC | |
| SBS | Chr8 | 21396007 | C-G | HET | INTERGENIC | |
| SBS | Chr8 | 5377270 | A-G | HET | INTERGENIC | |
| SBS | Chr8 | 5762893 | C-A | HET | INTERGENIC | |
| SBS | Chr8 | 7516616 | T-C | HOMO | INTERGENIC | |
| SBS | Chr9 | 16816762 | A-T | HOMO | INTERGENIC | |
| SBS | Chr9 | 19382949 | A-G | HET | INTERGENIC | |
| SBS | Chr9 | 20330377 | A-T | HET | UTR_3_PRIME | |
| SBS | Chr9 | 22594118 | G-A | HOMO | INTERGENIC |
Deletions: 32
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr3 | 649340 | 649348 | 8 | LOC_Os03g02040 |
| Deletion | Chr6 | 1550027 | 1550030 | 3 | |
| Deletion | Chr1 | 1564742 | 1564744 | 2 | LOC_Os01g03740 |
| Deletion | Chr6 | 1803642 | 1803643 | 1 | |
| Deletion | Chr2 | 1830779 | 1830780 | 1 | |
| Deletion | Chr5 | 1852687 | 1852691 | 4 | |
| Deletion | Chr1 | 2846423 | 2846425 | 2 | |
| Deletion | Chr7 | 3288736 | 3288737 | 1 | LOC_Os07g06740 |
| Deletion | Chr5 | 4287299 | 4287308 | 9 | |
| Deletion | Chr4 | 4691426 | 4691436 | 10 | |
| Deletion | Chr11 | 4788573 | 4788581 | 8 | LOC_Os11g09010 |
| Deletion | Chr10 | 5232045 | 5232047 | 2 | |
| Deletion | Chr4 | 5657366 | 5657367 | 1 | |
| Deletion | Chr6 | 14302284 | 14302285 | 1 | |
| Deletion | Chr7 | 14513856 | 14513859 | 3 | |
| Deletion | Chr7 | 14689240 | 14689248 | 8 | |
| Deletion | Chr9 | 15745605 | 15745607 | 2 | |
| Deletion | Chr11 | 16958187 | 16958188 | 1 | |
| Deletion | Chr12 | 20339543 | 20339546 | 3 | LOC_Os12g33690 |
| Deletion | Chr10 | 20616906 | 20616918 | 12 | LOC_Os10g38690 |
| Deletion | Chr9 | 21657901 | 21657903 | 2 | LOC_Os09g37580 |
| Deletion | Chr6 | 21882667 | 22079030 | 196363 | 28 |
| Deletion | Chr5 | 22693633 | 22693634 | 1 | |
| Deletion | Chr2 | 27426216 | 27427277 | 1061 | LOC_Os02g45180 |
| Deletion | Chr3 | 28382264 | 28382288 | 24 | |
| Deletion | Chr3 | 28787358 | 28787359 | 1 | |
| Deletion | Chr1 | 29711023 | 29711025 | 2 | |
| Deletion | Chr4 | 29778267 | 29778275 | 8 | LOC_Os04g49930 |
| Deletion | Chr4 | 31857468 | 31857475 | 7 | LOC_Os04g53496 |
| Deletion | Chr4 | 34901963 | 34901964 | 1 | LOC_Os04g58700 |
| Deletion | Chr1 | 38109618 | 38110909 | 1291 | |
| Deletion | Chr1 | 38547866 | 38549191 | 1325 |
Insertions: 18
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr7 | 159893 | 223105 | |
| Inversion | Chr2 | 24959736 | 24993385 | 2 |
No Translocation
