Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN-202 [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN-202 Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 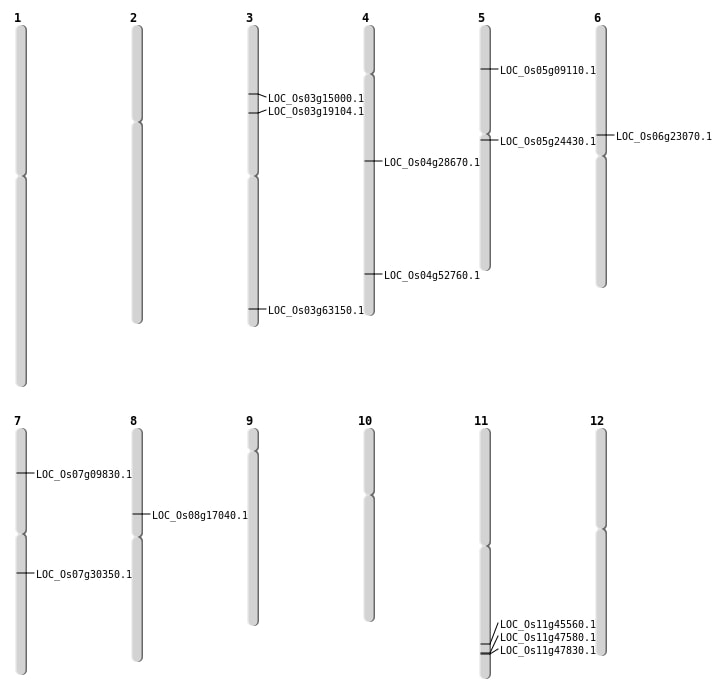
|
| Variant Information |
|---|
Single base substitutions: 39
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 18846146 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 2054824 | C-A | HOMO | INTRON | |
| SBS | Chr1 | 4812020 | C-G | HET | INTERGENIC | |
| SBS | Chr10 | 1125867 | G-T | HET | INTERGENIC | |
| SBS | Chr11 | 11586236 | T-C | HET | INTRON | |
| SBS | Chr11 | 14936758 | G-C | HET | INTERGENIC | |
| SBS | Chr12 | 15804124 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 1669130 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 19058134 | A-G | HET | INTRON | |
| SBS | Chr12 | 23805461 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr12 | 25862261 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr2 | 1018191 | A-G | HET | INTRON | |
| SBS | Chr2 | 20164573 | G-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr2 | 4461470 | C-T | HOMO | INTRON | |
| SBS | Chr3 | 10704768 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g19104 |
| SBS | Chr3 | 2648742 | C-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 2648743 | C-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 32415503 | T-C | HOMO | INTERGENIC | |
| SBS | Chr3 | 3908433 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 14634163 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 17468756 | G-T | HET | INTERGENIC | |
| SBS | Chr4 | 23058873 | G-T | HET | INTERGENIC | |
| SBS | Chr4 | 23070062 | C-A | HET | INTERGENIC | |
| SBS | Chr4 | 31377416 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 31413523 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g52760 |
| SBS | Chr5 | 17407639 | A-T | HOMO | INTERGENIC | |
| SBS | Chr5 | 20102967 | G-T | HOMO | UTR_3_PRIME | |
| SBS | Chr5 | 24118464 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr5 | 5074170 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os05g09110 |
| SBS | Chr5 | 5738708 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 21770702 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 5496827 | C-A | HOMO | INTERGENIC | |
| SBS | Chr7 | 17963923 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os07g30350 |
| SBS | Chr7 | 23128665 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 3281463 | G-A | HET | INTERGENIC | |
| SBS | Chr8 | 10435060 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os08g17040 |
| SBS | Chr8 | 4390024 | T-C | HET | INTERGENIC | |
| SBS | Chr9 | 7827552 | T-C | HET | INTERGENIC | |
| SBS | Chr9 | 8696998 | T-G | HET | INTRON |
Deletions: 22
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr4 | 1472987 | 1472988 | 1 | |
| Deletion | Chr4 | 1503323 | 1503324 | 1 | |
| Deletion | Chr7 | 5228596 | 5231977 | 3381 | LOC_Os07g09830 |
| Deletion | Chr6 | 8910997 | 8911002 | 5 | |
| Deletion | Chr6 | 11319399 | 11319403 | 4 | |
| Deletion | Chr11 | 11773410 | 11773416 | 6 | |
| Deletion | Chr1 | 13217488 | 13217496 | 8 | |
| Deletion | Chr6 | 13450582 | 13450585 | 3 | LOC_Os06g23070 |
| Deletion | Chr9 | 13831361 | 13831379 | 18 | |
| Deletion | Chr5 | 14107603 | 14107604 | 1 | LOC_Os05g24430 |
| Deletion | Chr3 | 16023274 | 16023275 | 1 | |
| Deletion | Chr4 | 16984531 | 16984532 | 1 | LOC_Os04g28670 |
| Deletion | Chr11 | 17369517 | 17369521 | 4 | |
| Deletion | Chr7 | 17905528 | 17905529 | 1 | |
| Deletion | Chr7 | 19026030 | 19026032 | 2 | |
| Deletion | Chr4 | 19779755 | 19779758 | 3 | |
| Deletion | Chr8 | 22317912 | 22317919 | 7 | |
| Deletion | Chr1 | 23277827 | 23277839 | 12 | |
| Deletion | Chr7 | 28897808 | 28897822 | 14 | |
| Deletion | Chr3 | 35694799 | 35694800 | 1 | LOC_Os03g63150 |
| Deletion | Chr3 | 35809429 | 35809443 | 14 | |
| Deletion | Chr1 | 37411025 | 37411032 | 7 |
Insertions: 6
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr10 | 2585702 | 2585704 | 3 | |
| Insertion | Chr11 | 2188507 | 2188507 | 1 | |
| Insertion | Chr11 | 28824595 | 28824596 | 2 | |
| Insertion | Chr3 | 8186693 | 8186695 | 3 | LOC_Os03g15000 |
| Insertion | Chr6 | 28570986 | 28570986 | 1 | |
| Insertion | Chr8 | 21075346 | 21075347 | 2 |
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr11 | 27579143 | 28852790 | 2 |
| Inversion | Chr11 | 28754912 | 28852779 | 2 |
Translocations: 1
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr9 | 13612984 | Chr1 | 35351937 |
