Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN-222 [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN-222 Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 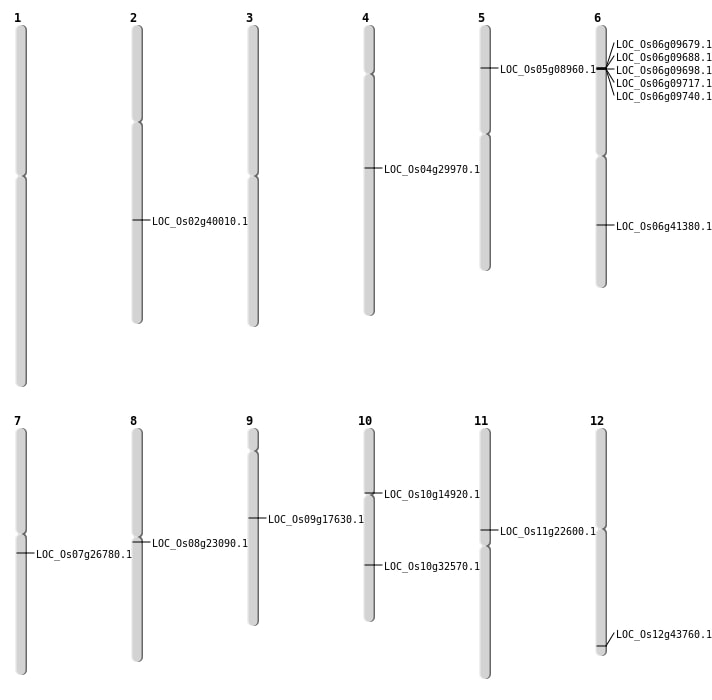
|
| Variant Information |
|---|
Single base substitutions: 41
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 13222706 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr1 | 14954797 | G-A | HOMO | INTERGENIC | |
| SBS | Chr1 | 16261932 | A-G | HOMO | INTRON | |
| SBS | Chr1 | 16568656 | C-T | HET | INTRON | |
| SBS | Chr1 | 26082720 | A-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 28510210 | T-C | HOMO | INTERGENIC | |
| SBS | Chr1 | 28611426 | G-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 28611427 | C-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 35990253 | T-C | HET | INTERGENIC | |
| SBS | Chr10 | 1208198 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 21834744 | T-C | HET | INTERGENIC | |
| SBS | Chr11 | 12998748 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os11g22600 |
| SBS | Chr11 | 3160972 | T-A | HET | INTRON | |
| SBS | Chr12 | 13218327 | A-C | HET | INTERGENIC | |
| SBS | Chr12 | 27186702 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g43760 |
| SBS | Chr12 | 3371955 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 3689731 | A-T | HET | INTERGENIC | |
| SBS | Chr12 | 5456273 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr3 | 27621966 | T-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 27622061 | C-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 30050196 | A-T | HET | INTERGENIC | |
| SBS | Chr3 | 32943044 | G-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 9420561 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 10679282 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 12731786 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 12926974 | A-C | HET | INTERGENIC | |
| SBS | Chr4 | 17882013 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g29970 |
| SBS | Chr4 | 18865297 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 6684702 | C-T | HET | INTRON | |
| SBS | Chr6 | 17621071 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 1771530 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 21605303 | T-A | HET | INTERGENIC | |
| SBS | Chr6 | 23337243 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 6442007 | A-T | HET | INTRON | |
| SBS | Chr7 | 13036449 | T-A | HET | INTRON | |
| SBS | Chr7 | 3977917 | G-T | HET | INTERGENIC | |
| SBS | Chr8 | 13909689 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os08g23090 |
| SBS | Chr8 | 13909690 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os08g23090 |
| SBS | Chr8 | 18249104 | C-T | HOMO | UTR_3_PRIME | |
| SBS | Chr8 | 6796294 | G-A | HET | INTRON | |
| SBS | Chr9 | 866309 | G-A | HOMO | SYNONYMOUS_CODING |
Deletions: 27
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr10 | 1210372 | 1210373 | 1 | |
| Deletion | Chr8 | 1476158 | 1476159 | 1 | |
| Deletion | Chr10 | 3128188 | 3128192 | 4 | |
| Deletion | Chr11 | 3593319 | 3593324 | 5 | |
| Deletion | Chr4 | 4249401 | 4249411 | 10 | |
| Deletion | Chr8 | 4866154 | 4866156 | 2 | |
| Deletion | Chr6 | 4935001 | 4965000 | 30000 | 5 |
| Deletion | Chr5 | 4958399 | 4958402 | 3 | LOC_Os05g08960 |
| Deletion | Chr6 | 6042371 | 6042373 | 2 | |
| Deletion | Chr5 | 7040429 | 7040431 | 2 | |
| Deletion | Chr5 | 7373054 | 7373056 | 2 | |
| Deletion | Chr10 | 7812599 | 7813851 | 1252 | LOC_Os10g14920 |
| Deletion | Chr11 | 8879379 | 8879389 | 10 | |
| Deletion | Chr1 | 10763994 | 10764000 | 6 | |
| Deletion | Chr9 | 10791548 | 10791561 | 13 | LOC_Os09g17630 |
| Deletion | Chr12 | 12612451 | 12612452 | 1 | |
| Deletion | Chr11 | 14309180 | 14309187 | 7 | |
| Deletion | Chr7 | 15473246 | 15473312 | 66 | LOC_Os07g26780 |
| Deletion | Chr9 | 15903196 | 15903199 | 3 | |
| Deletion | Chr10 | 17063944 | 17063953 | 9 | LOC_Os10g32570 |
| Deletion | Chr3 | 19283771 | 19283783 | 12 | |
| Deletion | Chr6 | 24808144 | 24808146 | 2 | LOC_Os06g41380 |
| Deletion | Chr6 | 25527885 | 25527899 | 14 | |
| Deletion | Chr3 | 25716243 | 25716244 | 1 | |
| Deletion | Chr4 | 27782038 | 27782039 | 1 | |
| Deletion | Chr7 | 27888478 | 27888518 | 40 | |
| Deletion | Chr1 | 28521296 | 28521305 | 9 |
Insertions: 27
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 24524866 | 24524869 | 4 | |
| Insertion | Chr1 | 3105924 | 3105926 | 3 | |
| Insertion | Chr10 | 17844109 | 17844109 | 1 | |
| Insertion | Chr10 | 19038169 | 19038172 | 4 | |
| Insertion | Chr11 | 19603304 | 19603306 | 3 | |
| Insertion | Chr11 | 19881096 | 19881097 | 2 | |
| Insertion | Chr11 | 4877628 | 4877644 | 17 | |
| Insertion | Chr11 | 4878367 | 4878368 | 2 | |
| Insertion | Chr12 | 25556821 | 25556828 | 8 | |
| Insertion | Chr12 | 4106207 | 4106207 | 1 | |
| Insertion | Chr12 | 6624869 | 6624870 | 2 | |
| Insertion | Chr2 | 24128220 | 24128223 | 4 | |
| Insertion | Chr2 | 24228585 | 24228590 | 6 | LOC_Os02g40010 |
| Insertion | Chr4 | 23881492 | 23881495 | 4 | |
| Insertion | Chr5 | 19546902 | 19546902 | 1 | |
| Insertion | Chr5 | 25234605 | 25234608 | 4 | |
| Insertion | Chr5 | 526189 | 526190 | 2 | |
| Insertion | Chr6 | 27130855 | 27130855 | 1 | |
| Insertion | Chr7 | 28895204 | 28895221 | 18 | |
| Insertion | Chr8 | 10188635 | 10188646 | 12 | |
| Insertion | Chr8 | 14740268 | 14740273 | 6 | |
| Insertion | Chr8 | 16701441 | 16701442 | 2 | |
| Insertion | Chr8 | 20204391 | 20204403 | 13 | |
| Insertion | Chr8 | 26805875 | 26805878 | 4 | |
| Insertion | Chr8 | 26815767 | 26815768 | 2 | |
| Insertion | Chr8 | 9879765 | 9879766 | 2 | |
| Insertion | Chr9 | 5093086 | 5093087 | 2 |
No Inversion
No Translocation
