Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN-223 [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN-223 Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 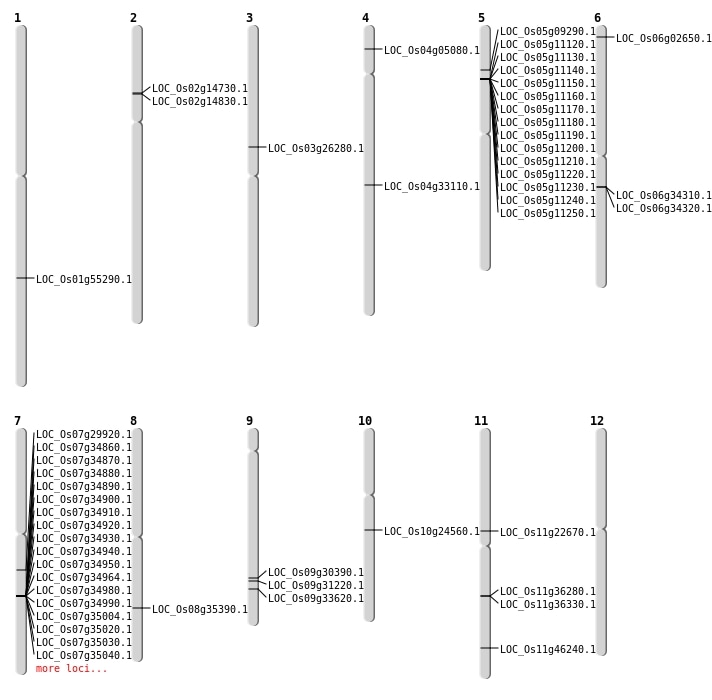
|
| Variant Information |
|---|
Single base substitutions: 38
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 4825528 | T-C | HOMO | INTERGENIC | |
| SBS | Chr10 | 11456677 | C-G | HET | INTRON | |
| SBS | Chr10 | 12604490 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os10g24560 |
| SBS | Chr10 | 21486934 | C-T | HOMO | UTR_3_PRIME | |
| SBS | Chr10 | 3305615 | T-C | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr11 | 12725366 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 28002736 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os11g46240 |
| SBS | Chr11 | 4503285 | T-C | HET | INTERGENIC | |
| SBS | Chr2 | 11632002 | A-C | HET | INTERGENIC | |
| SBS | Chr2 | 25722705 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 25873727 | A-T | HET | INTERGENIC | |
| SBS | Chr2 | 8241437 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os02g14830 |
| SBS | Chr2 | 9227930 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 12417477 | A-T | HET | INTERGENIC | |
| SBS | Chr3 | 15029788 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os03g26280 |
| SBS | Chr3 | 30377951 | C-G | HOMO | INTERGENIC | |
| SBS | Chr3 | 30793571 | C-T | HET | INTRON | |
| SBS | Chr3 | 7326975 | A-C | HET | INTRON | |
| SBS | Chr4 | 29034416 | C-T | HET | INTRON | |
| SBS | Chr5 | 19219343 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 21971406 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 5190180 | C-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os05g09290 |
| SBS | Chr6 | 17740080 | G-C | HET | INTERGENIC | |
| SBS | Chr6 | 31023554 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 4773081 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 994685 | T-C | HET | INTERGENIC | |
| SBS | Chr7 | 119652 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 17611430 | C-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os07g29920 |
| SBS | Chr8 | 21573894 | A-C | HET | INTRON | |
| SBS | Chr8 | 22311258 | T-C | HOMO | NON_SYNONYMOUS_CODING | LOC_Os08g35390 |
| SBS | Chr8 | 8303236 | C-A | HOMO | INTRON | |
| SBS | Chr9 | 18781316 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os09g31220 |
| SBS | Chr9 | 18781317 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os09g31220 |
| SBS | Chr9 | 19660402 | A-G | HET | INTERGENIC | |
| SBS | Chr9 | 19856807 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os09g33620 |
| SBS | Chr9 | 19856808 | A-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr9 | 19856814 | A-T | HET | STOP_GAINED | LOC_Os09g33620 |
| SBS | Chr9 | 21617295 | G-C | HET | INTERGENIC |
Deletions: 29
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr6 | 942113 | 947182 | 5069 | LOC_Os06g02650 |
| Deletion | Chr5 | 2017677 | 2017679 | 2 | |
| Deletion | Chr4 | 2514403 | 2514404 | 1 | LOC_Os04g05080 |
| Deletion | Chr4 | 2835636 | 2835637 | 1 | |
| Deletion | Chr8 | 2904402 | 2904406 | 4 | |
| Deletion | Chr12 | 4538343 | 4538344 | 1 | |
| Deletion | Chr5 | 6246001 | 6358000 | 112000 | 14 |
| Deletion | Chr2 | 11147109 | 11147110 | 1 | |
| Deletion | Chr11 | 13040987 | 13040991 | 4 | LOC_Os11g22670 |
| Deletion | Chr9 | 13809814 | 13809815 | 1 | |
| Deletion | Chr6 | 13839340 | 13839341 | 1 | |
| Deletion | Chr6 | 16940951 | 16940955 | 4 | |
| Deletion | Chr6 | 17746817 | 17746835 | 18 | |
| Deletion | Chr5 | 17831417 | 17831418 | 1 | |
| Deletion | Chr9 | 18496186 | 18496189 | 3 | LOC_Os09g30390 |
| Deletion | Chr10 | 19611468 | 19611469 | 1 | |
| Deletion | Chr6 | 19959614 | 19959616 | 2 | 2 |
| Deletion | Chr9 | 20871389 | 20871392 | 3 | |
| Deletion | Chr7 | 20887001 | 21311000 | 424000 | 72 |
| Deletion | Chr2 | 23843011 | 23843013 | 2 | |
| Deletion | Chr7 | 24406077 | 24406082 | 5 | |
| Deletion | Chr2 | 25533710 | 25533719 | 9 | |
| Deletion | Chr2 | 25748713 | 25748725 | 12 | |
| Deletion | Chr2 | 25927370 | 25927373 | 3 | |
| Deletion | Chr12 | 26267631 | 26267633 | 2 | |
| Deletion | Chr7 | 28257919 | 28257934 | 15 | |
| Deletion | Chr6 | 29586295 | 29586297 | 2 | |
| Deletion | Chr3 | 31073836 | 31073838 | 2 | |
| Deletion | Chr1 | 31826898 | 31826899 | 1 | LOC_Os01g55290 |
Insertions: 17
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 3318561 | 3318564 | 4 | |
| Insertion | Chr1 | 9391364 | 9391375 | 12 | |
| Insertion | Chr10 | 2238013 | 2238014 | 2 | |
| Insertion | Chr10 | 2674547 | 2674548 | 2 | |
| Insertion | Chr12 | 21282427 | 21282428 | 2 | |
| Insertion | Chr12 | 27134457 | 27134460 | 4 | |
| Insertion | Chr12 | 9328593 | 9328594 | 2 | |
| Insertion | Chr2 | 18453732 | 18453739 | 8 | |
| Insertion | Chr2 | 19219927 | 19219930 | 4 | |
| Insertion | Chr2 | 24146279 | 24146282 | 4 | |
| Insertion | Chr3 | 11684668 | 11684671 | 4 | |
| Insertion | Chr3 | 7620255 | 7620256 | 2 | |
| Insertion | Chr4 | 20046112 | 20046112 | 1 | LOC_Os04g33110 |
| Insertion | Chr7 | 3643477 | 3643479 | 3 | |
| Insertion | Chr8 | 13595894 | 13595894 | 1 | |
| Insertion | Chr8 | 16843436 | 16843437 | 2 | |
| Insertion | Chr8 | 18371379 | 18371380 | 2 |
Inversions: 8
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr5 | 4630060 | 6245587 | |
| Inversion | Chr5 | 4630077 | 6375602 | |
| Inversion | Chr5 | 5700068 | 5736018 | |
| Inversion | Chr5 | 5700087 | 5736035 | |
| Inversion | Chr2 | 6901411 | 8136603 | LOC_Os02g14730 |
| Inversion | Chr2 | 6901431 | 8136611 | LOC_Os02g14730 |
| Inversion | Chr11 | 21367256 | 21402891 | LOC_Os11g36280 |
| Inversion | Chr11 | 21370720 | 21405713 | LOC_Os11g36330 |
Translocations: 6
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr6 | 946178 | Chr2 | 2070469 | LOC_Os06g02650 |
| Translocation | Chr6 | 947179 | Chr2 | 2070472 | |
| Translocation | Chr11 | 21367291 | Chr4 | 5013927 | LOC_Os11g36280 |
| Translocation | Chr11 | 21370711 | Chr4 | 5013931 | |
| Translocation | Chr7 | 29233976 | Chr6 | 942126 | LOC_Os06g02650 |
| Translocation | Chr7 | 29234002 | Chr6 | 946114 | LOC_Os06g02650 |
