Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN-228 [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN-228 Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 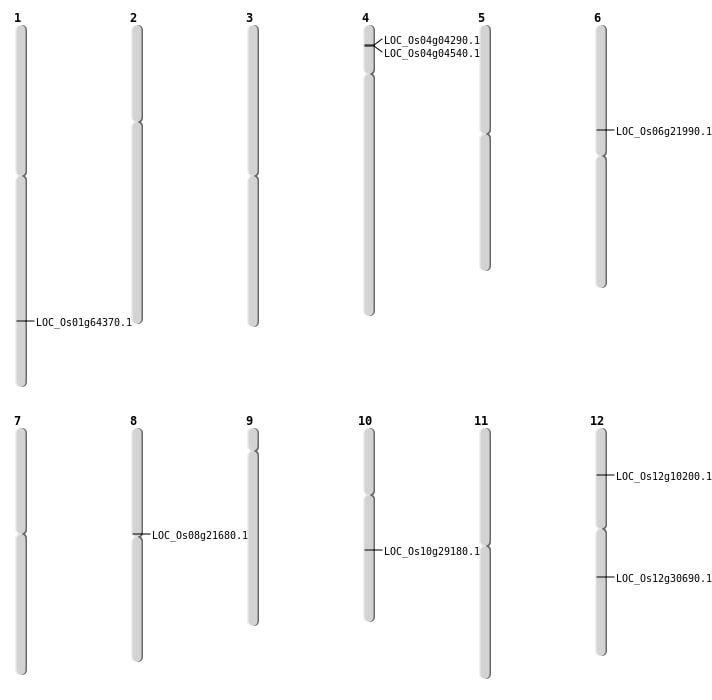
|
| Variant Information |
|---|
Single base substitutions: 29
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 10563766 | A-G | HET | INTRON | |
| SBS | Chr1 | 17787185 | A-T | HET | INTERGENIC | |
| SBS | Chr1 | 20603868 | G-T | HET | INTERGENIC | |
| SBS | Chr1 | 32755922 | A-G | HET | INTRON | |
| SBS | Chr10 | 13274191 | T-A | HET | INTERGENIC | |
| SBS | Chr10 | 15201043 | A-G | HET | UTR_3_PRIME | |
| SBS | Chr10 | 16056175 | A-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr11 | 11962215 | T-A | HET | INTRON | |
| SBS | Chr11 | 19175080 | T-C | HET | INTERGENIC | |
| SBS | Chr12 | 26254693 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 15475597 | G-T | HOMO | INTRON | |
| SBS | Chr2 | 21515964 | C-G | HET | INTERGENIC | |
| SBS | Chr2 | 34614182 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr2 | 8957402 | C-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 24085890 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 2007181 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g04290 |
| SBS | Chr4 | 23297364 | A-T | HET | INTRON | |
| SBS | Chr4 | 29375153 | A-C | HOMO | INTERGENIC | |
| SBS | Chr5 | 16143458 | C-T | HOMO | INTRON | |
| SBS | Chr5 | 8124108 | G-A | HOMO | INTRON | |
| SBS | Chr6 | 3743842 | A-G | HET | INTERGENIC | |
| SBS | Chr6 | 8819816 | A-G | HET | INTERGENIC | |
| SBS | Chr7 | 22105537 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 24033496 | G-A | HOMO | INTERGENIC | |
| SBS | Chr7 | 28083487 | T-A | HOMO | INTERGENIC | |
| SBS | Chr7 | 8154681 | C-A | HET | INTERGENIC | |
| SBS | Chr8 | 12917824 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os08g21680 |
| SBS | Chr9 | 16695657 | A-G | HET | INTERGENIC | |
| SBS | Chr9 | 6735634 | G-C | HET | INTERGENIC |
Deletions: 15
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr10 | 1586953 | 1586965 | 12 | |
| Deletion | Chr2 | 4070964 | 4070966 | 2 | |
| Deletion | Chr12 | 5392152 | 5392173 | 21 | LOC_Os12g10200 |
| Deletion | Chr7 | 8151676 | 8151681 | 5 | |
| Deletion | Chr1 | 9113208 | 9113216 | 8 | |
| Deletion | Chr7 | 11082075 | 11082082 | 7 | |
| Deletion | Chr6 | 12719236 | 12719244 | 8 | LOC_Os06g21990 |
| Deletion | Chr10 | 15201046 | 15201047 | 1 | LOC_Os10g29180 |
| Deletion | Chr5 | 16050013 | 16050017 | 4 | |
| Deletion | Chr1 | 17811110 | 17812643 | 1533 | |
| Deletion | Chr9 | 19442269 | 19442270 | 1 | |
| Deletion | Chr9 | 21126526 | 21126540 | 14 | |
| Deletion | Chr1 | 26162775 | 26162776 | 1 | |
| Deletion | Chr3 | 26333692 | 26333693 | 1 | |
| Deletion | Chr1 | 37360549 | 37360552 | 3 | LOC_Os01g64370 |
No Insertion
Inversions: 4
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr4 | 2172389 | 5326992 | LOC_Os04g04540 |
| Inversion | Chr8 | 3566404 | 4417000 | |
| Inversion | Chr1 | 15900796 | 15920108 | |
| Inversion | Chr12 | 18409827 | 18430286 | LOC_Os12g30690 |
No Translocation
