Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN-231 [Download] |
| Generation | M2-SOS |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN-231 Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 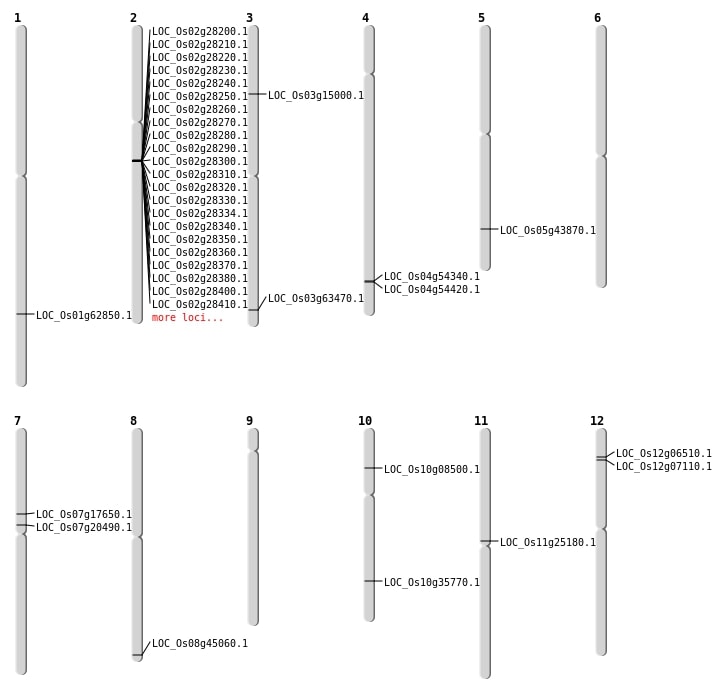
|
| Variant Information |
|---|
Single base substitutions: 41
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 36046113 | C-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 5492806 | G-T | HOMO | INTERGENIC | |
| SBS | Chr10 | 15468601 | C-G | HOMO | INTERGENIC | |
| SBS | Chr10 | 4612441 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os10g08500 |
| SBS | Chr10 | 4612442 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os10g08500 |
| SBS | Chr11 | 14347148 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os11g25180 |
| SBS | Chr11 | 23761178 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 24402534 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 4546172 | G-A | HOMO | INTRON | |
| SBS | Chr11 | 813916 | A-G | HET | UTR_5_PRIME | |
| SBS | Chr12 | 3148499 | A-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os12g06510 |
| SBS | Chr12 | 4962219 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 24181089 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os02g39990 |
| SBS | Chr2 | 24181090 | C-A | HET | STOP_GAINED | LOC_Os02g39990 |
| SBS | Chr2 | 26304188 | T-G | HOMO | INTERGENIC | |
| SBS | Chr2 | 3320695 | C-A | HET | INTERGENIC | |
| SBS | Chr2 | 8044308 | T-C | HET | INTRON | |
| SBS | Chr2 | 9105659 | A-G | HET | INTERGENIC | |
| SBS | Chr3 | 11494658 | C-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 14591058 | G-T | HET | INTERGENIC | |
| SBS | Chr3 | 24675741 | T-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 2553157 | C-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 35859190 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g63470 |
| SBS | Chr3 | 35859191 | A-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr3 | 5546678 | C-T | HOMO | INTERGENIC | |
| SBS | Chr4 | 21766876 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 23624265 | G-T | HET | INTERGENIC | |
| SBS | Chr4 | 32347307 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g54340 |
| SBS | Chr4 | 4433987 | C-A | HET | INTERGENIC | |
| SBS | Chr5 | 8664270 | C-T | HOMO | INTERGENIC | |
| SBS | Chr6 | 16808642 | G-C | HET | INTERGENIC | |
| SBS | Chr6 | 23803414 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 9697005 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 11260739 | C-A | HET | INTERGENIC | |
| SBS | Chr7 | 19044128 | T-C | HET | INTERGENIC | |
| SBS | Chr7 | 19156138 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 16804135 | G-A | HET | INTERGENIC | |
| SBS | Chr8 | 24532370 | A-C | HET | INTERGENIC | |
| SBS | Chr9 | 16081181 | T-C | HET | INTRON | |
| SBS | Chr9 | 16883188 | T-A | HET | INTERGENIC | |
| SBS | Chr9 | 4399123 | C-T | HOMO | INTERGENIC |
Deletions: 30
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr4 | 700500 | 700512 | 12 | |
| Deletion | Chr10 | 1804277 | 1804278 | 1 | |
| Deletion | Chr1 | 1885989 | 1886001 | 12 | |
| Deletion | Chr2 | 3652697 | 3652704 | 7 | |
| Deletion | Chr2 | 4621336 | 4621343 | 7 | |
| Deletion | Chr5 | 5590108 | 5590128 | 20 | |
| Deletion | Chr10 | 7264602 | 7264610 | 8 | |
| Deletion | Chr1 | 7583842 | 7583845 | 3 | |
| Deletion | Chr9 | 8641905 | 8641907 | 2 | |
| Deletion | Chr1 | 9803752 | 9803753 | 1 | |
| Deletion | Chr3 | 10657379 | 10657380 | 1 | |
| Deletion | Chr2 | 11601177 | 11601182 | 5 | |
| Deletion | Chr7 | 11832094 | 11832103 | 9 | LOC_Os07g20490 |
| Deletion | Chr9 | 12038716 | 12038718 | 2 | |
| Deletion | Chr1 | 12167208 | 12167209 | 1 | |
| Deletion | Chr2 | 12623127 | 12623145 | 18 | |
| Deletion | Chr3 | 16163163 | 16163164 | 1 | |
| Deletion | Chr2 | 16688001 | 17337000 | 649000 | 97 |
| Deletion | Chr2 | 21236066 | 21236081 | 15 | LOC_Os02g35315 |
| Deletion | Chr4 | 22832705 | 22832706 | 1 | |
| Deletion | Chr1 | 22885533 | 22885539 | 6 | |
| Deletion | Chr7 | 23069448 | 23069465 | 17 | |
| Deletion | Chr4 | 23313653 | 23313655 | 2 | |
| Deletion | Chr5 | 25509479 | 25509482 | 3 | LOC_Os05g43870 |
| Deletion | Chr1 | 26845765 | 26845766 | 1 | |
| Deletion | Chr6 | 27256071 | 27256075 | 4 | |
| Deletion | Chr8 | 28290140 | 28290149 | 9 | LOC_Os08g45060 |
| Deletion | Chr3 | 28564253 | 28564255 | 2 | |
| Deletion | Chr5 | 28768691 | 28768692 | 1 | |
| Deletion | Chr1 | 36399215 | 36399224 | 9 | LOC_Os01g62850 |
Insertions: 21
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 11768578 | 11768579 | 2 | |
| Insertion | Chr1 | 33401530 | 33401531 | 2 | |
| Insertion | Chr1 | 9445153 | 9445154 | 2 | |
| Insertion | Chr10 | 17838567 | 17838570 | 4 | |
| Insertion | Chr10 | 19133335 | 19133340 | 6 | LOC_Os10g35770 |
| Insertion | Chr11 | 1709375 | 1709375 | 1 | |
| Insertion | Chr11 | 19595026 | 19595027 | 2 | |
| Insertion | Chr11 | 28694685 | 28694686 | 2 | |
| Insertion | Chr11 | 28980609 | 28980614 | 6 | |
| Insertion | Chr12 | 24620953 | 24620970 | 18 | |
| Insertion | Chr12 | 3489260 | 3489260 | 1 | LOC_Os12g07110 |
| Insertion | Chr12 | 9389281 | 9389284 | 4 | |
| Insertion | Chr2 | 8450981 | 8450982 | 2 | |
| Insertion | Chr3 | 1257473 | 1257474 | 2 | |
| Insertion | Chr3 | 8186693 | 8186695 | 3 | LOC_Os03g15000 |
| Insertion | Chr4 | 32381953 | 32381961 | 9 | LOC_Os04g54420 |
| Insertion | Chr5 | 29132340 | 29132340 | 1 | |
| Insertion | Chr6 | 20677302 | 20677303 | 2 | |
| Insertion | Chr6 | 28902624 | 28902624 | 1 | |
| Insertion | Chr7 | 4014182 | 4014183 | 2 | |
| Insertion | Chr7 | 4510894 | 4510895 | 2 |
Inversions: 3
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr7 | 9588202 | 10426729 | LOC_Os07g17650 |
| Inversion | Chr3 | 23972520 | 30355667 | |
| Inversion | Chr3 | 23972546 | 30355683 |
No Translocation
