Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN-234 [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN-234 Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 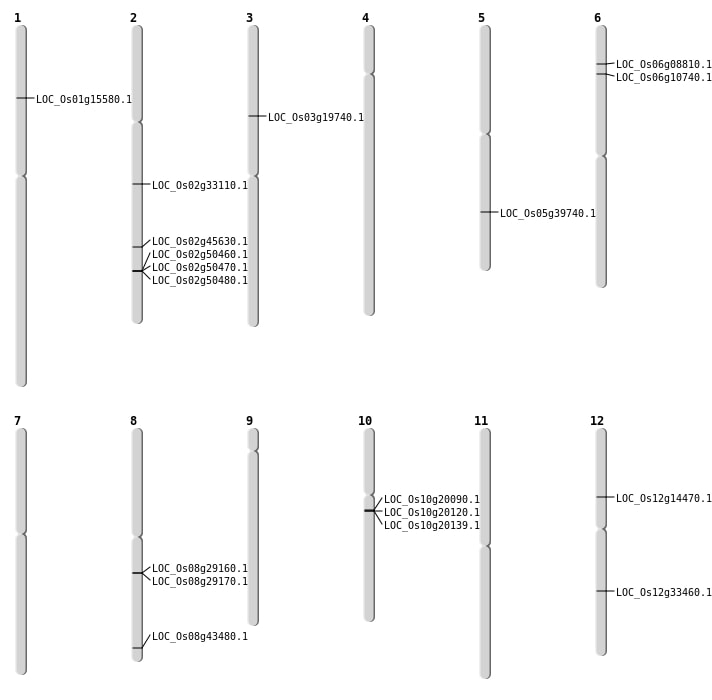
|
| Variant Information |
|---|
Single base substitutions: 33
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 30701995 | G-T | HET | INTERGENIC | |
| SBS | Chr1 | 34226345 | C-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 36337939 | C-A | HET | INTERGENIC | |
| SBS | Chr1 | 36930410 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 10562304 | T-A | HET | INTERGENIC | |
| SBS | Chr10 | 13150706 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 16227292 | A-C | HET | INTRON | |
| SBS | Chr10 | 16423707 | A-T | HET | INTERGENIC | |
| SBS | Chr10 | 16423710 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 20220439 | T-G | HET | NON_SYNONYMOUS_CODING | LOC_Os12g33460 |
| SBS | Chr12 | 8546455 | C-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 23163661 | T-C | HOMO | INTERGENIC | |
| SBS | Chr2 | 3850729 | T-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 3850869 | A-G | HOMO | INTERGENIC | |
| SBS | Chr3 | 22394952 | C-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 29104159 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 31960318 | G-A | HET | INTRON | |
| SBS | Chr4 | 17682770 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 5810741 | C-A | HOMO | UTR_3_PRIME | |
| SBS | Chr4 | 5810755 | A-G | HOMO | UTR_3_PRIME | |
| SBS | Chr5 | 10081935 | C-T | HOMO | INTERGENIC | |
| SBS | Chr5 | 1704313 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 25089931 | A-T | HOMO | INTERGENIC | |
| SBS | Chr6 | 11894333 | T-A | HET | INTERGENIC | |
| SBS | Chr6 | 1650930 | T-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr7 | 5282975 | A-T | HET | INTERGENIC | |
| SBS | Chr7 | 5635559 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 13553796 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 21269762 | A-G | HOMO | INTERGENIC | |
| SBS | Chr8 | 23529978 | G-T | HOMO | INTERGENIC | |
| SBS | Chr8 | 27784487 | C-T | HOMO | INTRON | |
| SBS | Chr8 | 8683162 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr9 | 2172131 | A-G | HET | INTERGENIC |
Deletions: 28
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr1 | 241582 | 241583 | 1 | |
| Deletion | Chr4 | 2043339 | 2043340 | 1 | |
| Deletion | Chr5 | 3654518 | 3654522 | 4 | |
| Deletion | Chr1 | 4112195 | 4112415 | 220 | |
| Deletion | Chr6 | 5608401 | 5611417 | 3016 | LOC_Os06g10740 |
| Deletion | Chr4 | 5870486 | 5870494 | 8 | |
| Deletion | Chr9 | 5910866 | 5910870 | 4 | |
| Deletion | Chr7 | 8370737 | 8370738 | 1 | |
| Deletion | Chr2 | 9609367 | 9609377 | 10 | |
| Deletion | Chr12 | 9672411 | 9672454 | 43 | |
| Deletion | Chr10 | 10092843 | 10110785 | 17942 | 3 |
| Deletion | Chr10 | 10093001 | 10115000 | 22000 | 3 |
| Deletion | Chr9 | 10770669 | 10770678 | 9 | |
| Deletion | Chr3 | 11097682 | 11097684 | 2 | LOC_Os03g19740 |
| Deletion | Chr5 | 13426072 | 13426073 | 1 | |
| Deletion | Chr8 | 17857637 | 17857640 | 3 | 2 |
| Deletion | Chr10 | 18469956 | 18469957 | 1 | |
| Deletion | Chr10 | 19515831 | 19515833 | 2 | |
| Deletion | Chr2 | 19684963 | 19684969 | 6 | LOC_Os02g33110 |
| Deletion | Chr4 | 20336413 | 20336421 | 8 | |
| Deletion | Chr4 | 20666687 | 20666688 | 1 | |
| Deletion | Chr4 | 21454333 | 21454377 | 44 | |
| Deletion | Chr5 | 22687858 | 22687860 | 2 | |
| Deletion | Chr5 | 23322758 | 23341776 | 19018 | LOC_Os05g39740 |
| Deletion | Chr6 | 24193531 | 24193539 | 8 | |
| Deletion | Chr4 | 26499813 | 26499814 | 1 | |
| Deletion | Chr2 | 30802817 | 30824891 | 22074 | 3 |
| Deletion | Chr1 | 33173054 | 33173063 | 9 |
Insertions: 1
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr8 | 27503156 | 27503158 | 3 | LOC_Os08g43480 |
Inversions: 9
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr1 | 4112205 | 8751081 | LOC_Os01g15580 |
| Inversion | Chr1 | 4112411 | 8751079 | LOC_Os01g15580 |
| Inversion | Chr6 | 4405791 | 5916768 | LOC_Os06g08810 |
| Inversion | Chr12 | 8255431 | 8258742 | LOC_Os12g14470 |
| Inversion | Chr12 | 8258743 | 8259241 | |
| Inversion | Chr10 | 13149108 | 13262381 | |
| Inversion | Chr10 | 13149137 | 13262385 | |
| Inversion | Chr12 | 13379536 | 13403669 | |
| Inversion | Chr2 | 27757258 | 27814772 | LOC_Os02g45630 |
Translocations: 1
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr9 | 3070109 | Chr6 | 5608401 |
