Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN-235 [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN-235 Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 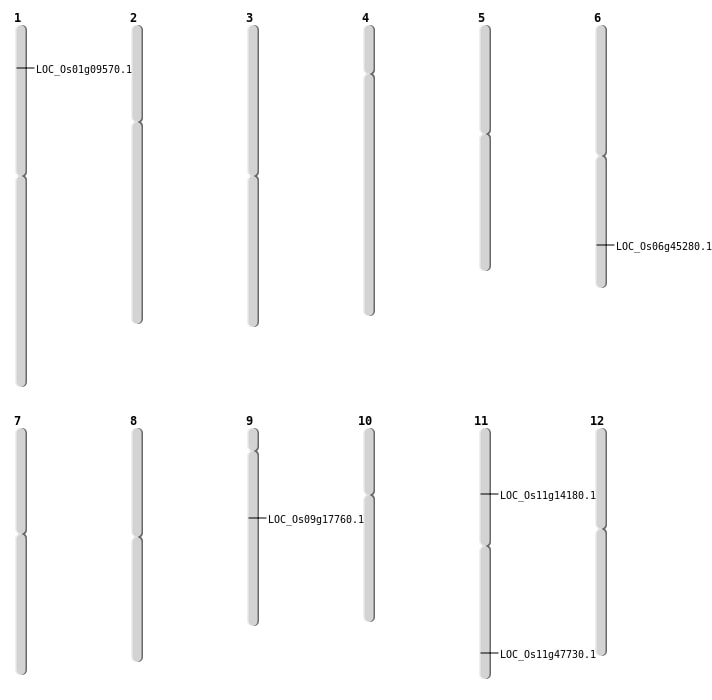
|
| Variant Information |
|---|
Single base substitutions: 32
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 1431727 | C-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 1431729 | G-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 37905411 | G-T | HET | INTRON | |
| SBS | Chr1 | 39197333 | C-A | HET | INTERGENIC | |
| SBS | Chr10 | 18430982 | A-T | HET | INTERGENIC | |
| SBS | Chr10 | 2267999 | G-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr11 | 7936815 | A-G | HET | INTERGENIC | |
| SBS | Chr11 | 9034685 | A-T | HET | INTRON | |
| SBS | Chr12 | 26586218 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 26606627 | A-G | HET | INTERGENIC | |
| SBS | Chr12 | 461361 | A-T | HET | INTERGENIC | |
| SBS | Chr12 | 9657317 | A-G | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr3 | 29437026 | A-G | HOMO | INTERGENIC | |
| SBS | Chr3 | 29438823 | C-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 29438824 | G-T | HOMO | INTERGENIC | |
| SBS | Chr4 | 33190407 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 7588897 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 17790378 | A-C | HET | INTRON | |
| SBS | Chr5 | 17947904 | T-A | HET | INTERGENIC | |
| SBS | Chr6 | 5947138 | G-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 12687366 | G-T | HET | INTERGENIC | |
| SBS | Chr7 | 18550731 | T-G | HET | INTERGENIC | |
| SBS | Chr8 | 10922145 | A-G | HET | INTERGENIC | |
| SBS | Chr8 | 12640991 | A-G | HET | INTERGENIC | |
| SBS | Chr8 | 1966973 | C-A | HET | INTERGENIC | |
| SBS | Chr8 | 22736485 | A-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr8 | 23667308 | A-T | HOMO | INTERGENIC | |
| SBS | Chr8 | 28320224 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 12025861 | T-A | HET | INTERGENIC | |
| SBS | Chr9 | 12069378 | G-T | HET | INTRON | |
| SBS | Chr9 | 12069379 | A-T | HET | INTRON | |
| SBS | Chr9 | 12096901 | T-A | HET | INTERGENIC |
Deletions: 24
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr8 | 1956064 | 1956065 | 1 | |
| Deletion | Chr10 | 2235054 | 2235055 | 1 | |
| Deletion | Chr1 | 4909288 | 4909291 | 3 | LOC_Os01g09570 |
| Deletion | Chr11 | 7927895 | 7927908 | 13 | |
| Deletion | Chr11 | 7934974 | 7934976 | 2 | LOC_Os11g14180 |
| Deletion | Chr9 | 11942929 | 11942930 | 1 | |
| Deletion | Chr8 | 11966998 | 11966999 | 1 | |
| Deletion | Chr5 | 12392399 | 12392400 | 1 | |
| Deletion | Chr1 | 12980353 | 12980357 | 4 | |
| Deletion | Chr11 | 15504954 | 15504955 | 1 | |
| Deletion | Chr6 | 17884130 | 17884139 | 9 | |
| Deletion | Chr8 | 18122360 | 18122362 | 2 | |
| Deletion | Chr10 | 18236596 | 18236597 | 1 | |
| Deletion | Chr4 | 20162537 | 20162538 | 1 | |
| Deletion | Chr5 | 21829116 | 21829118 | 2 | |
| Deletion | Chr12 | 22876765 | 22876766 | 1 | |
| Deletion | Chr1 | 23413567 | 23413568 | 1 | |
| Deletion | Chr4 | 26187860 | 26194813 | 6953 | |
| Deletion | Chr6 | 26303714 | 26303720 | 6 | |
| Deletion | Chr1 | 26845557 | 26845559 | 2 | |
| Deletion | Chr2 | 27368304 | 27368307 | 3 | |
| Deletion | Chr6 | 27369219 | 27369224 | 5 | LOC_Os06g45280 |
| Deletion | Chr1 | 30331552 | 30331553 | 1 | |
| Deletion | Chr1 | 30466354 | 30466358 | 4 |
Insertions: 18
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 14632859 | 14632862 | 4 | |
| Insertion | Chr1 | 8938808 | 8938810 | 3 | |
| Insertion | Chr11 | 18673792 | 18673794 | 3 | |
| Insertion | Chr11 | 22168333 | 22168334 | 2 | |
| Insertion | Chr11 | 28794547 | 28794554 | 8 | LOC_Os11g47730 |
| Insertion | Chr12 | 23060486 | 23060486 | 1 | |
| Insertion | Chr12 | 23060491 | 23060491 | 1 | |
| Insertion | Chr2 | 16683162 | 16683162 | 1 | |
| Insertion | Chr2 | 18910158 | 18910175 | 18 | |
| Insertion | Chr2 | 19293462 | 19293463 | 2 | |
| Insertion | Chr2 | 24120906 | 24120907 | 2 | |
| Insertion | Chr2 | 24416504 | 24416505 | 2 | |
| Insertion | Chr2 | 29867953 | 29867956 | 4 | |
| Insertion | Chr4 | 27932931 | 27932931 | 1 | |
| Insertion | Chr6 | 9678593 | 9678594 | 2 | |
| Insertion | Chr7 | 10920496 | 10920497 | 2 | |
| Insertion | Chr8 | 15238874 | 15238875 | 2 | |
| Insertion | Chr8 | 8336123 | 8336124 | 2 |
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr9 | 10857019 | 12072337 | LOC_Os09g17760 |
No Translocation
Tandem duplications: 1
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Tandem Duplication | Chr4 | 3962998 | 26194794 | 22231796 |
