Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN-246 [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN-246 Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 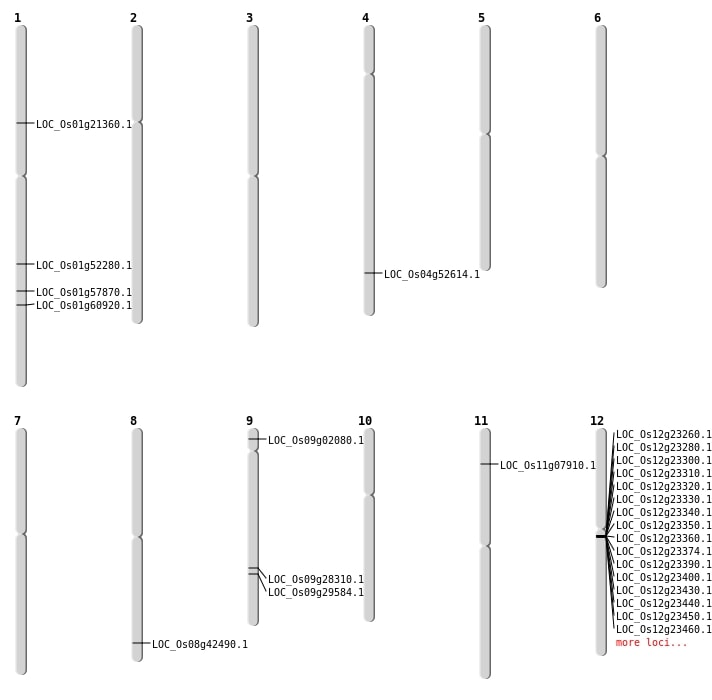
|
| Variant Information |
|---|
Single base substitutions: 40
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 1604415 | A-T | HOMO | UTR_3_PRIME | |
| SBS | Chr1 | 31337583 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 31337584 | T-A | HET | INTERGENIC | |
| SBS | Chr1 | 31709282 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 32743063 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 33463059 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os01g57870 |
| SBS | Chr1 | 34311629 | A-G | HET | INTRON | |
| SBS | Chr1 | 34311638 | G-A | HET | INTRON | |
| SBS | Chr1 | 41788282 | C-T | HET | INTRON | |
| SBS | Chr1 | 43024685 | A-T | HET | INTERGENIC | |
| SBS | Chr10 | 16407846 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr10 | 20788262 | G-A | HOMO | INTERGENIC | |
| SBS | Chr11 | 22034211 | C-T | HOMO | INTERGENIC | |
| SBS | Chr11 | 25632500 | A-G | HET | INTERGENIC | |
| SBS | Chr12 | 19963046 | G-T | HET | INTERGENIC | |
| SBS | Chr12 | 8985179 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 10441273 | T-A | HET | INTERGENIC | |
| SBS | Chr2 | 20748897 | T-G | HET | INTRON | |
| SBS | Chr2 | 27042432 | A-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 3707259 | T-A | HET | INTRON | |
| SBS | Chr3 | 24688379 | G-C | HET | INTERGENIC | |
| SBS | Chr3 | 26326096 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr3 | 27887062 | T-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 4467224 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 5465129 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 10981049 | C-T | HET | INTRON | |
| SBS | Chr5 | 25552838 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 25552846 | A-T | HET | INTERGENIC | |
| SBS | Chr5 | 26248921 | A-T | HET | INTERGENIC | |
| SBS | Chr5 | 26376512 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 16536956 | A-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 22089512 | C-T | HOMO | INTRON | |
| SBS | Chr8 | 11885003 | A-T | HOMO | INTERGENIC | |
| SBS | Chr8 | 15318632 | C-T | HET | INTRON | |
| SBS | Chr8 | 22935189 | A-G | HET | INTERGENIC | |
| SBS | Chr8 | 6711300 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 13937965 | A-G | HET | INTERGENIC | |
| SBS | Chr9 | 19029062 | G-A | HET | INTRON | |
| SBS | Chr9 | 6759114 | G-C | HET | INTERGENIC | |
| SBS | Chr9 | 764939 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os09g02080 |
Deletions: 19
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr11 | 1622333 | 1622342 | 9 | |
| Deletion | Chr9 | 1782762 | 1782780 | 18 | |
| Deletion | Chr11 | 2432391 | 2432399 | 8 | |
| Deletion | Chr11 | 4082054 | 4082055 | 1 | LOC_Os11g07910 |
| Deletion | Chr2 | 8118816 | 8118822 | 6 | |
| Deletion | Chr5 | 11643581 | 11643585 | 4 | |
| Deletion | Chr1 | 11936796 | 11936802 | 6 | LOC_Os01g21360 |
| Deletion | Chr4 | 12289468 | 12289469 | 1 | |
| Deletion | Chr12 | 13155001 | 13574000 | 419000 | 55 |
| Deletion | Chr2 | 18731747 | 18731766 | 19 | |
| Deletion | Chr1 | 19990079 | 19990080 | 1 | |
| Deletion | Chr12 | 26158362 | 26158372 | 10 | |
| Deletion | Chr8 | 26862676 | 26862695 | 19 | LOC_Os08g42490 |
| Deletion | Chr1 | 30056721 | 30056735 | 14 | LOC_Os01g52280 |
| Deletion | Chr4 | 31299831 | 31299876 | 45 | LOC_Os04g52614 |
| Deletion | Chr3 | 33844421 | 33844426 | 5 | |
| Deletion | Chr1 | 34225520 | 34225521 | 1 | |
| Deletion | Chr1 | 35238563 | 35240298 | 1735 | LOC_Os01g60920 |
| Deletion | Chr1 | 36005981 | 36005983 | 2 |
Insertions: 3
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr10 | 20317590 | 20317590 | 1 | |
| Insertion | Chr10 | 21857128 | 21857131 | 4 | |
| Insertion | Chr11 | 13209863 | 13209863 | 1 |
Inversions: 5
Translocations: 9
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 10368080 | Chr3 | 11289216 | LOC_Os11g18366 |
| Translocation | Chr11 | 10368104 | Chr3 | 11291416 | LOC_Os11g18366 |
| Translocation | Chr9 | 17192153 | Chr6 | 6805724 | LOC_Os09g28310 |
| Translocation | Chr9 | 17192159 | Chr6 | 6808456 | LOC_Os09g28310 |
| Translocation | Chr9 | 17209655 | Chr6 | 5778580 | |
| Translocation | Chr9 | 17209670 | Chr3 | 27808037 | |
| Translocation | Chr9 | 17210656 | Chr3 | 27808026 | |
| Translocation | Chr9 | 17963356 | Chr5 | 20175405 | |
| Translocation | Chr9 | 17992550 | Chr5 | 20175507 | LOC_Os09g29584 |
