Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN-274 [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN-274 Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 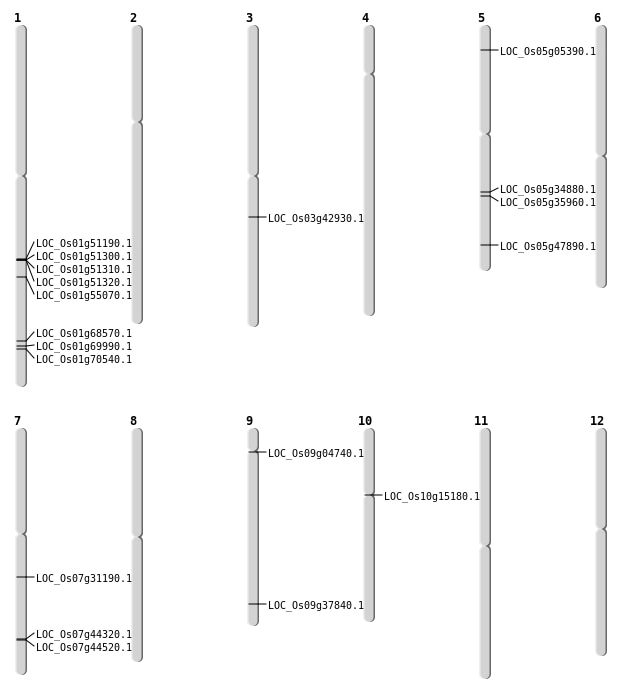
|
| Variant Information |
|---|
Single base substitutions: 38
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 1276263 | T-G | HET | INTERGENIC | |
| SBS | Chr1 | 19201939 | T-A | HET | INTERGENIC | |
| SBS | Chr1 | 21307269 | G-A | HOMO | INTRON | |
| SBS | Chr1 | 40488205 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os01g69990 |
| SBS | Chr1 | 40488206 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os01g69990 |
| SBS | Chr10 | 4499147 | T-C | HOMO | INTRON | |
| SBS | Chr11 | 14729663 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 16191804 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 7806242 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 10608311 | T-G | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr12 | 12610525 | G-T | HOMO | INTRON | |
| SBS | Chr12 | 26718513 | A-G | HET | INTERGENIC | |
| SBS | Chr2 | 3343332 | A-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 25753389 | C-G | HET | INTRON | |
| SBS | Chr3 | 26209529 | C-T | HET | INTRON | |
| SBS | Chr3 | 30729365 | G-A | HET | INTRON | |
| SBS | Chr4 | 22484090 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 1018805 | G-C | HET | INTERGENIC | |
| SBS | Chr5 | 14129105 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 16133376 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 16133377 | C-A | HET | INTERGENIC | |
| SBS | Chr5 | 1682074 | C-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr5 | 21312318 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os05g35960 |
| SBS | Chr6 | 20454048 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 29806092 | G-A | HOMO | INTERGENIC | |
| SBS | Chr6 | 30582379 | T-G | HET | INTRON | |
| SBS | Chr7 | 16554116 | C-A | HET | INTRON | |
| SBS | Chr7 | 26349033 | C-T | HET | INTRON | |
| SBS | Chr7 | 26475507 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os07g44320 |
| SBS | Chr7 | 26585213 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os07g44520 |
| SBS | Chr7 | 26585218 | T-G | HET | NON_SYNONYMOUS_CODING | LOC_Os07g44520 |
| SBS | Chr7 | 8885360 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 12848467 | T-C | HOMO | INTRON | |
| SBS | Chr8 | 18323289 | G-A | HET | INTRON | |
| SBS | Chr8 | 20309935 | C-T | HOMO | INTERGENIC | |
| SBS | Chr8 | 21262826 | T-C | HOMO | INTERGENIC | |
| SBS | Chr8 | 28403398 | T-A | HET | INTRON | |
| SBS | Chr9 | 2526641 | T-G | HET | NON_SYNONYMOUS_CODING | LOC_Os09g04740 |
Deletions: 21
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr5 | 2678167 | 2678169 | 2 | LOC_Os05g05390 |
| Deletion | Chr5 | 2678173 | 2678175 | 2 | LOC_Os05g05390 |
| Deletion | Chr1 | 13029840 | 13029865 | 25 | |
| Deletion | Chr9 | 14202343 | 14202346 | 3 | |
| Deletion | Chr10 | 14771805 | 14771813 | 8 | |
| Deletion | Chr7 | 18462219 | 18462228 | 9 | LOC_Os07g31190 |
| Deletion | Chr3 | 18822392 | 18822396 | 4 | |
| Deletion | Chr3 | 20165639 | 20165641 | 2 | |
| Deletion | Chr4 | 21798759 | 21798770 | 11 | |
| Deletion | Chr6 | 21817983 | 21817986 | 3 | |
| Deletion | Chr5 | 22136475 | 22136490 | 15 | |
| Deletion | Chr5 | 22732748 | 22732753 | 5 | |
| Deletion | Chr3 | 23942933 | 23942934 | 1 | LOC_Os03g42930 |
| Deletion | Chr4 | 27285735 | 27285736 | 1 | |
| Deletion | Chr5 | 27461055 | 27461060 | 5 | LOC_Os05g47890 |
| Deletion | Chr2 | 28175254 | 28175285 | 31 | |
| Deletion | Chr1 | 28610468 | 28610474 | 6 | |
| Deletion | Chr1 | 29501001 | 29517000 | 16000 | 3 |
| Deletion | Chr1 | 31661957 | 31661962 | 5 | LOC_Os01g55070 |
| Deletion | Chr1 | 39830485 | 39830494 | 9 | LOC_Os01g68570 |
| Deletion | Chr1 | 40856729 | 40856737 | 8 | LOC_Os01g70540 |
Insertions: 7
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr1 | 29425343 | 29516339 | LOC_Os01g51190 |
Translocations: 3
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 13737708 | Chr10 | 8063297 | LOC_Os10g15180 |
| Translocation | Chr5 | 20725568 | Chr2 | 5558377 | LOC_Os05g34880 |
| Translocation | Chr9 | 21809955 | Chr4 | 22380836 | LOC_Os09g37840 |
