Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN-277 [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN-277 Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 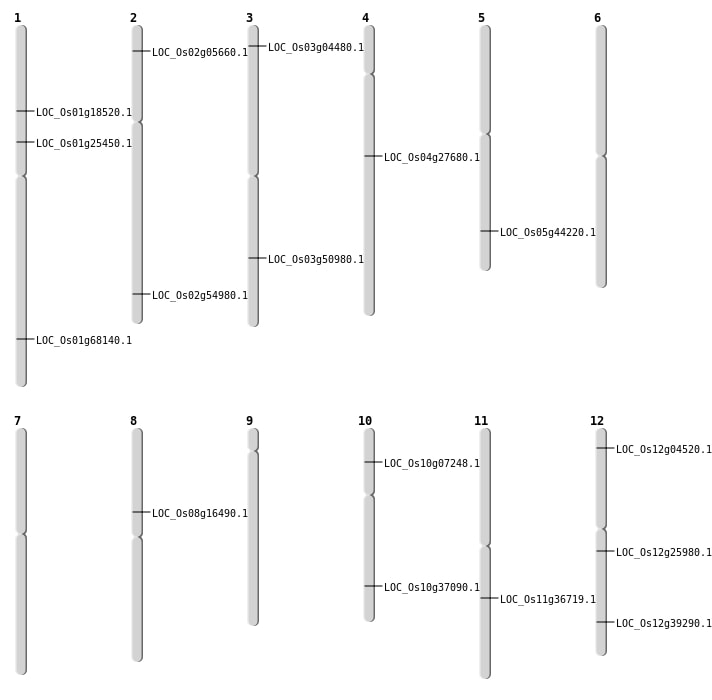
|
| Variant Information |
|---|
Single base substitutions: 29
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 12514939 | G-T | HET | INTRON | |
| SBS | Chr1 | 28124056 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 15043871 | C-T | HOMO | INTERGENIC | |
| SBS | Chr10 | 22967845 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 3818259 | A-G | HOMO | NON_SYNONYMOUS_CODING | LOC_Os10g07248 |
| SBS | Chr11 | 10845900 | C-T | HET | INTRON | |
| SBS | Chr11 | 13893192 | T-C | HET | INTRON | |
| SBS | Chr11 | 16954827 | T-C | HET | INTERGENIC | |
| SBS | Chr11 | 21682420 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os11g36719 |
| SBS | Chr11 | 26698469 | T-C | HET | INTERGENIC | |
| SBS | Chr12 | 15080919 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os12g25980 |
| SBS | Chr12 | 17264618 | C-A | HET | INTRON | |
| SBS | Chr12 | 1926022 | C-T | HET | SPLICE_SITE_DONOR | LOC_Os12g04520 |
| SBS | Chr12 | 26986387 | G-A | HOMO | INTERGENIC | |
| SBS | Chr12 | 27207326 | C-A | HOMO | INTERGENIC | |
| SBS | Chr12 | 4572535 | T-C | HET | INTERGENIC | |
| SBS | Chr2 | 30277452 | G-T | HOMO | INTRON | |
| SBS | Chr2 | 32108794 | C-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 15525151 | G-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 10842004 | A-G | HET | INTRON | |
| SBS | Chr4 | 19950274 | G-T | HOMO | INTERGENIC | |
| SBS | Chr4 | 24344997 | C-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 34563303 | T-A | HET | INTERGENIC | |
| SBS | Chr5 | 13509023 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 1275066 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 11070203 | C-T | HOMO | INTRON | |
| SBS | Chr7 | 5532845 | T-C | HOMO | INTERGENIC | |
| SBS | Chr7 | 6894587 | C-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 7075499 | A-T | HET | INTERGENIC |
Deletions: 30
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr6 | 1118471 | 1118472 | 1 | |
| Deletion | Chr5 | 1979420 | 1979426 | 6 | |
| Deletion | Chr3 | 2075735 | 2075741 | 6 | LOC_Os03g04480 |
| Deletion | Chr10 | 2233698 | 2233730 | 32 | |
| Deletion | Chr8 | 2421557 | 2421559 | 2 | |
| Deletion | Chr2 | 2766342 | 2766355 | 13 | LOC_Os02g05660 |
| Deletion | Chr1 | 3929820 | 3929830 | 10 | |
| Deletion | Chr11 | 5540213 | 5540214 | 1 | |
| Deletion | Chr3 | 6252983 | 6252987 | 4 | |
| Deletion | Chr5 | 6481585 | 6481586 | 1 | |
| Deletion | Chr5 | 8753870 | 8753876 | 6 | |
| Deletion | Chr1 | 9115496 | 9115497 | 1 | |
| Deletion | Chr5 | 10097204 | 10097225 | 21 | |
| Deletion | Chr1 | 10417208 | 10417219 | 11 | LOC_Os01g18520 |
| Deletion | Chr11 | 13685347 | 13685348 | 1 | |
| Deletion | Chr1 | 14419612 | 14419620 | 8 | LOC_Os01g25450 |
| Deletion | Chr11 | 14937244 | 14937246 | 2 | |
| Deletion | Chr10 | 16490895 | 16490899 | 4 | |
| Deletion | Chr10 | 19854509 | 19854524 | 15 | LOC_Os10g37090 |
| Deletion | Chr2 | 20365767 | 20365796 | 29 | |
| Deletion | Chr8 | 21154098 | 21154099 | 1 | |
| Deletion | Chr5 | 21597947 | 21597953 | 6 | |
| Deletion | Chr1 | 22196347 | 22196361 | 14 | |
| Deletion | Chr1 | 23423597 | 23423700 | 103 | |
| Deletion | Chr4 | 24345332 | 24345333 | 1 | |
| Deletion | Chr2 | 24865291 | 24865315 | 24 | |
| Deletion | Chr5 | 25727951 | 25727956 | 5 | LOC_Os05g44220 |
| Deletion | Chr6 | 25908738 | 25908739 | 1 | |
| Deletion | Chr2 | 33688821 | 33688843 | 22 | LOC_Os02g54980 |
| Deletion | Chr1 | 39614297 | 39614306 | 9 | LOC_Os01g68140 |
Insertions: 10
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr1 | 16919640 | 17353819 |
Translocations: 7
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr8 | 3468692 | Chr1 | 17353846 | |
| Translocation | Chr8 | 3468695 | Chr1 | 17353849 | |
| Translocation | Chr8 | 10108691 | Chr6 | 18097955 | LOC_Os08g16490 |
| Translocation | Chr4 | 16311351 | Chr1 | 34334168 | |
| Translocation | Chr4 | 16363771 | Chr1 | 23524840 | LOC_Os04g27680 |
| Translocation | Chr12 | 24178281 | Chr3 | 29127918 | 2 |
| Translocation | Chr12 | 24178282 | Chr3 | 29130497 | 2 |
