Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN-278 [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN-278 Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 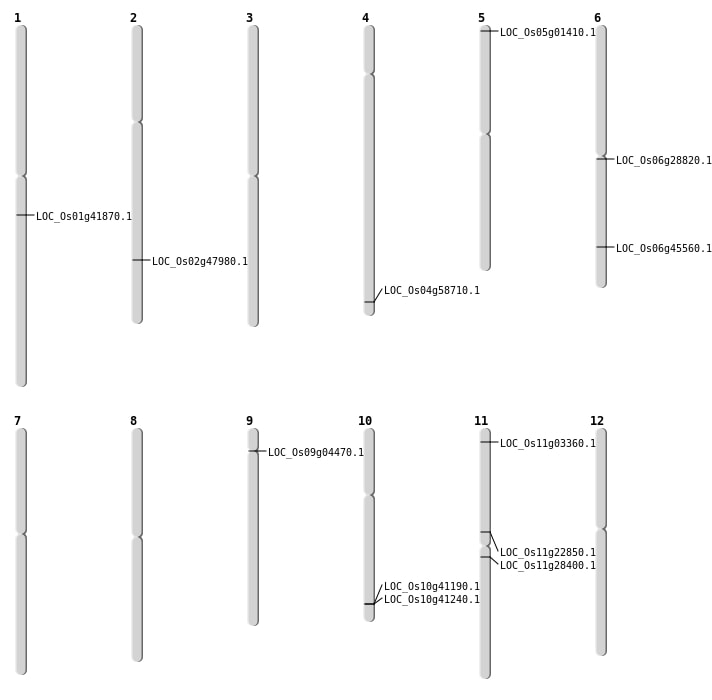
|
| Variant Information |
|---|
Single base substitutions: 46
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 1448524 | A-G | HET | SPLICE_SITE_REGION | |
| SBS | Chr1 | 18254458 | C-T | HET | INTRON | |
| SBS | Chr1 | 26418162 | G-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 7106171 | T-A | HET | INTRON | |
| SBS | Chr1 | 7106173 | T-A | HET | INTRON | |
| SBS | Chr10 | 499474 | C-A | HET | INTERGENIC | |
| SBS | Chr11 | 1173029 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 1267401 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os11g03360 |
| SBS | Chr11 | 1267402 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os11g03360 |
| SBS | Chr11 | 13143512 | T-G | HET | NON_SYNONYMOUS_CODING | LOC_Os11g22850 |
| SBS | Chr11 | 13527008 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 16347208 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os11g28400 |
| SBS | Chr11 | 20494484 | T-A | HET | INTERGENIC | |
| SBS | Chr11 | 3455944 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr12 | 17333924 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 19808219 | A-G | HOMO | UTR_3_PRIME | |
| SBS | Chr12 | 19808277 | C-T | HOMO | UTR_3_PRIME | |
| SBS | Chr12 | 25170572 | G-A | HET | INTRON | |
| SBS | Chr2 | 13945668 | G-C | HET | INTERGENIC | |
| SBS | Chr2 | 4203124 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 7608222 | G-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 8047764 | C-T | HET | INTRON | |
| SBS | Chr3 | 17954274 | G-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 25296654 | C-A | HET | INTERGENIC | |
| SBS | Chr3 | 33686502 | G-A | HOMO | INTRON | |
| SBS | Chr4 | 12169523 | T-A | HET | INTERGENIC | |
| SBS | Chr4 | 7357479 | T-C | HET | INTRON | |
| SBS | Chr4 | 9617658 | T-G | HET | INTERGENIC | |
| SBS | Chr5 | 19521954 | T-G | HET | INTERGENIC | |
| SBS | Chr5 | 20228414 | T-A | HET | INTRON | |
| SBS | Chr5 | 241623 | G-A | HET | STOP_GAINED | LOC_Os05g01410 |
| SBS | Chr5 | 7292286 | G-T | HET | INTERGENIC | |
| SBS | Chr5 | 9823778 | C-T | HET | INTRON | |
| SBS | Chr6 | 11320121 | C-T | HOMO | INTERGENIC | |
| SBS | Chr6 | 17364850 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 20136991 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 27584608 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os06g45560 |
| SBS | Chr6 | 29380150 | G-T | HET | INTERGENIC | |
| SBS | Chr6 | 3737863 | A-G | HET | SPLICE_SITE_REGION | |
| SBS | Chr7 | 2164815 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr8 | 18182441 | A-G | HOMO | INTERGENIC | |
| SBS | Chr8 | 21650572 | T-C | HOMO | INTERGENIC | |
| SBS | Chr8 | 27746761 | C-T | HOMO | INTERGENIC | |
| SBS | Chr8 | 5787131 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 6083102 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 2208603 | T-C | HOMO | INTRON |
Deletions: 26
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr10 | 499527 | 499569 | 42 | |
| Deletion | Chr9 | 1566118 | 1566119 | 1 | |
| Deletion | Chr9 | 1852685 | 1852693 | 8 | |
| Deletion | Chr9 | 2382261 | 2382262 | 1 | LOC_Os09g04470 |
| Deletion | Chr5 | 5371300 | 5371308 | 8 | |
| Deletion | Chr5 | 6599677 | 6599680 | 3 | |
| Deletion | Chr11 | 7591046 | 7592956 | 1910 | |
| Deletion | Chr2 | 7608197 | 7608215 | 18 | |
| Deletion | Chr6 | 11198889 | 11198895 | 6 | |
| Deletion | Chr12 | 13406645 | 13406670 | 25 | |
| Deletion | Chr7 | 13898461 | 13898463 | 2 | |
| Deletion | Chr6 | 16413237 | 16413242 | 5 | LOC_Os06g28820 |
| Deletion | Chr11 | 17818688 | 17818692 | 4 | |
| Deletion | Chr10 | 18320925 | 18320929 | 4 | |
| Deletion | Chr10 | 18991595 | 18991612 | 17 | |
| Deletion | Chr12 | 19808560 | 19808562 | 2 | |
| Deletion | Chr11 | 20295803 | 20295804 | 1 | |
| Deletion | Chr5 | 22547964 | 22547968 | 4 | |
| Deletion | Chr3 | 23061188 | 23061202 | 14 | |
| Deletion | Chr1 | 23730951 | 23731403 | 452 | LOC_Os01g41870 |
| Deletion | Chr5 | 24362226 | 24362227 | 1 | |
| Deletion | Chr1 | 25800579 | 25800580 | 1 | |
| Deletion | Chr7 | 29192113 | 29192117 | 4 | |
| Deletion | Chr1 | 29851215 | 29851216 | 1 | |
| Deletion | Chr1 | 29990673 | 29990682 | 9 | |
| Deletion | Chr4 | 34909982 | 34909992 | 10 | LOC_Os04g58710 |
Insertions: 7
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr10 | 11743742 | 11743745 | 4 | |
| Insertion | Chr11 | 23208034 | 23208036 | 3 | |
| Insertion | Chr11 | 3710388 | 3710389 | 2 | |
| Insertion | Chr2 | 18298484 | 18298489 | 6 | |
| Insertion | Chr2 | 29365387 | 29365389 | 3 | LOC_Os02g47980 |
| Insertion | Chr3 | 29291431 | 29291432 | 2 | |
| Insertion | Chr8 | 5657557 | 5657560 | 4 |
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr10 | 22124987 | 22161483 | 2 |
| Inversion | Chr10 | 22125001 | 22161486 | 2 |
Translocations: 5
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr7 | 7356401 | Chr1 | 39678865 | |
| Translocation | Chr11 | 9031430 | Chr7 | 23098259 | |
| Translocation | Chr11 | 9031545 | Chr7 | 23098251 | |
| Translocation | Chr12 | 10171784 | Chr2 | 17608408 | |
| Translocation | Chr6 | 24301539 | Chr2 | 25044490 |
