Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN-279 [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN-279 Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 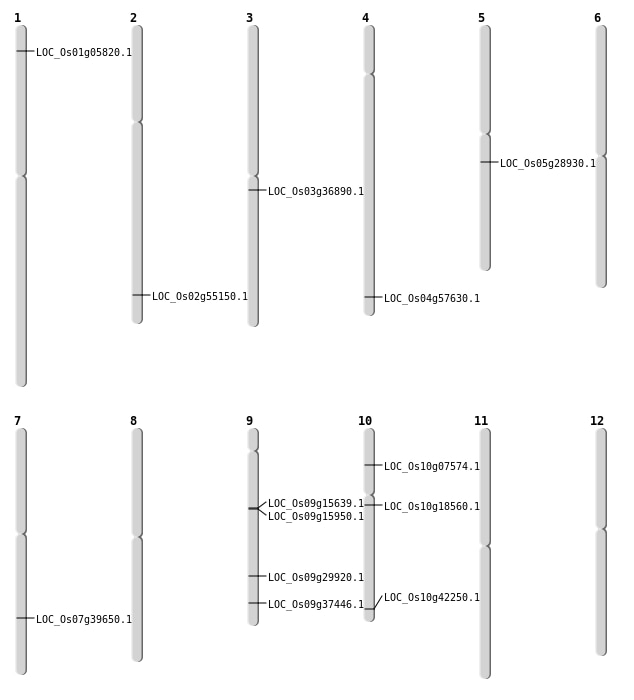
|
| Variant Information |
|---|
Single base substitutions: 35
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 26890903 | A-T | HOMO | INTRON | |
| SBS | Chr1 | 29573238 | T-C | HET | INTRON | |
| SBS | Chr1 | 32877970 | G-T | HET | UTR_3_PRIME | |
| SBS | Chr1 | 32877971 | C-T | HET | UTR_3_PRIME | |
| SBS | Chr1 | 32971142 | C-A | HET | INTRON | |
| SBS | Chr1 | 32971143 | T-A | HET | INTRON | |
| SBS | Chr1 | 41129440 | C-T | HOMO | INTERGENIC | |
| SBS | Chr10 | 14650975 | T-C | HET | INTERGENIC | |
| SBS | Chr10 | 17518130 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 18644280 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 22754221 | C-T | HET | START_GAINED | LOC_Os10g42250 |
| SBS | Chr10 | 4217907 | T-C | HOMO | NON_SYNONYMOUS_CODING | LOC_Os10g07574 |
| SBS | Chr10 | 4779901 | G-A | HET | INTRON | |
| SBS | Chr11 | 4811654 | C-A | HET | INTRON | |
| SBS | Chr2 | 25935044 | T-A | HET | INTERGENIC | |
| SBS | Chr2 | 28248407 | C-A | HET | INTERGENIC | |
| SBS | Chr2 | 7933905 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 20468485 | T-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os03g36890 |
| SBS | Chr3 | 28843336 | C-G | HET | INTRON | |
| SBS | Chr3 | 35633215 | A-G | HET | INTERGENIC | |
| SBS | Chr3 | 9105288 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 17074289 | T-C | HOMO | INTERGENIC | |
| SBS | Chr4 | 1857780 | A-G | HOMO | INTERGENIC | |
| SBS | Chr4 | 19097501 | A-G | HOMO | INTERGENIC | |
| SBS | Chr4 | 22264568 | G-T | HET | INTERGENIC | |
| SBS | Chr5 | 1285772 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 2580015 | C-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr6 | 9675605 | T-C | HET | INTERGENIC | |
| SBS | Chr7 | 19007593 | A-T | HET | INTERGENIC | |
| SBS | Chr8 | 20125684 | C-T | HOMO | INTERGENIC | |
| SBS | Chr9 | 18191703 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr9 | 18194137 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os09g29920 |
| SBS | Chr9 | 1923851 | A-T | HET | INTERGENIC | |
| SBS | Chr9 | 21596362 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os09g37446 |
| SBS | Chr9 | 9898663 | C-T | HOMO | INTERGENIC |
Deletions: 19
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr12 | 2576576 | 2576577 | 1 | |
| Deletion | Chr11 | 4173414 | 4173424 | 10 | |
| Deletion | Chr7 | 7069742 | 7069747 | 5 | |
| Deletion | Chr12 | 7459405 | 7459423 | 18 | |
| Deletion | Chr2 | 7945548 | 7945557 | 9 | |
| Deletion | Chr12 | 15612020 | 15612023 | 3 | |
| Deletion | Chr4 | 15810971 | 15810979 | 8 | |
| Deletion | Chr4 | 21621523 | 21621532 | 9 | |
| Deletion | Chr12 | 22445069 | 22445088 | 19 | |
| Deletion | Chr8 | 24985938 | 24985939 | 1 | |
| Deletion | Chr5 | 28229283 | 28229284 | 1 | |
| Deletion | Chr6 | 28485803 | 28485814 | 11 | |
| Deletion | Chr3 | 30416296 | 30416297 | 1 | |
| Deletion | Chr1 | 31878385 | 31878386 | 1 | |
| Deletion | Chr3 | 33692914 | 33692919 | 5 | |
| Deletion | Chr2 | 33786352 | 33786354 | 2 | LOC_Os02g55150 |
| Deletion | Chr4 | 34299923 | 34299924 | 1 | LOC_Os04g57630 |
| Deletion | Chr2 | 34397709 | 34397714 | 5 | |
| Deletion | Chr1 | 40420582 | 40420601 | 19 |
Insertions: 12
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 18409844 | 18409845 | 2 | |
| Insertion | Chr1 | 23572413 | 23572414 | 2 | |
| Insertion | Chr1 | 38599018 | 38599033 | 16 | |
| Insertion | Chr1 | 42965165 | 42965166 | 2 | |
| Insertion | Chr1 | 4915293 | 4915294 | 2 | |
| Insertion | Chr10 | 9413777 | 9413778 | 2 | LOC_Os10g18560 |
| Insertion | Chr3 | 11684668 | 11684671 | 4 | |
| Insertion | Chr5 | 29609065 | 29609065 | 1 | |
| Insertion | Chr5 | 4809099 | 4809101 | 3 | |
| Insertion | Chr6 | 12843205 | 12843206 | 2 | |
| Insertion | Chr6 | 23545576 | 23545578 | 3 | |
| Insertion | Chr9 | 10486654 | 10486655 | 2 |
No Inversion
Translocations: 10
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr9 | 9544628 | Chr4 | 12757392 | LOC_Os09g15639 |
| Translocation | Chr9 | 9746730 | Chr4 | 1012802 | LOC_Os09g15950 |
| Translocation | Chr9 | 18188815 | Chr5 | 16959731 | LOC_Os05g28930 |
| Translocation | Chr9 | 18188817 | Chr1 | 5623793 | |
| Translocation | Chr9 | 18195003 | Chr1 | 5623808 | |
| Translocation | Chr9 | 18195006 | Chr5 | 16959741 | LOC_Os05g28930 |
| Translocation | Chr11 | 18291043 | Chr8 | 15196565 | |
| Translocation | Chr8 | 19838638 | Chr6 | 7023925 | |
| Translocation | Chr7 | 23767471 | Chr3 | 25304104 | LOC_Os07g39650 |
| Translocation | Chr7 | 24498027 | Chr1 | 2784232 | LOC_Os01g05820 |
