Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN-281 [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN-281 Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 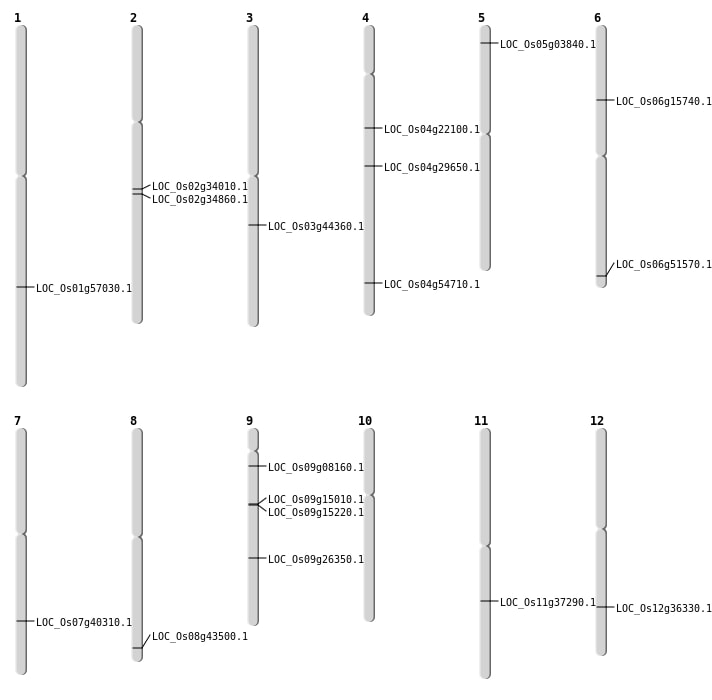
|
| Variant Information |
|---|
Single base substitutions: 43
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 10896279 | C-T | HET | INTRON | |
| SBS | Chr1 | 14846713 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 20682027 | T-C | HOMO | INTRON | |
| SBS | Chr1 | 24900146 | C-A | HET | INTRON | |
| SBS | Chr1 | 41157499 | T-G | HET | NON_SYNONYMOUS_CODING | LOC_Os01g71106 |
| SBS | Chr1 | 493054 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 493055 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 5174596 | T-A | HET | INTERGENIC | |
| SBS | Chr11 | 21247129 | T-A | HET | INTERGENIC | |
| SBS | Chr12 | 10141008 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 22246635 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os12g36330 |
| SBS | Chr2 | 26257654 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 28501472 | T-A | HET | INTERGENIC | |
| SBS | Chr2 | 3562564 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 11933779 | G-T | HET | INTERGENIC | |
| SBS | Chr3 | 12020620 | G-A | HET | INTRON | |
| SBS | Chr3 | 15495240 | T-A | HET | INTERGENIC | |
| SBS | Chr3 | 17438769 | C-T | HET | UTR_3_PRIME | |
| SBS | Chr3 | 24937269 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os03g44360 |
| SBS | Chr3 | 27255212 | A-T | HET | INTRON | |
| SBS | Chr3 | 35210359 | T-C | HET | INTRON | |
| SBS | Chr4 | 17645433 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os04g29650 |
| SBS | Chr4 | 24228385 | T-C | HOMO | INTERGENIC | |
| SBS | Chr4 | 35384625 | C-A | HET | UTR_3_PRIME | |
| SBS | Chr5 | 17761251 | C-A | HET | INTERGENIC | |
| SBS | Chr5 | 21921896 | A-G | HET | INTERGENIC | |
| SBS | Chr5 | 483047 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 19267263 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 20206778 | C-T | HOMO | SPLICE_SITE_REGION | |
| SBS | Chr6 | 3481468 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 7341541 | A-G | HET | INTERGENIC | |
| SBS | Chr7 | 2458484 | T-G | HOMO | UTR_3_PRIME | |
| SBS | Chr7 | 24786351 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 12133727 | A-T | HET | INTRON | |
| SBS | Chr8 | 15523497 | G-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr8 | 2101536 | T-G | HET | INTERGENIC | |
| SBS | Chr9 | 11090714 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 1334781 | T-C | HET | INTRON | |
| SBS | Chr9 | 15917548 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os09g26350 |
| SBS | Chr9 | 15917554 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os09g26350 |
| SBS | Chr9 | 18930572 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 4229133 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os09g08160 |
| SBS | Chr9 | 9039859 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os09g15010 |
Deletions: 20
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr5 | 1708547 | 1708552 | 5 | LOC_Os05g03840 |
| Deletion | Chr1 | 5403245 | 5403252 | 7 | |
| Deletion | Chr3 | 6058062 | 6058066 | 4 | |
| Deletion | Chr9 | 9238251 | 9238254 | 3 | LOC_Os09g15220 |
| Deletion | Chr8 | 10452271 | 10452273 | 2 | |
| Deletion | Chr10 | 10767712 | 10767717 | 5 | |
| Deletion | Chr11 | 11116746 | 11116750 | 4 | |
| Deletion | Chr8 | 12076851 | 12076854 | 3 | |
| Deletion | Chr5 | 13567123 | 13567126 | 3 | |
| Deletion | Chr1 | 13619936 | 13619942 | 6 | |
| Deletion | Chr4 | 18630688 | 18630691 | 3 | |
| Deletion | Chr9 | 20242200 | 20242201 | 1 | |
| Deletion | Chr8 | 20982481 | 20982493 | 12 | |
| Deletion | Chr7 | 24175331 | 24175332 | 1 | LOC_Os07g40310 |
| Deletion | Chr8 | 24407571 | 24407588 | 17 | |
| Deletion | Chr2 | 24880567 | 24880575 | 8 | |
| Deletion | Chr1 | 29443377 | 29443378 | 1 | |
| Deletion | Chr3 | 29810001 | 29821000 | 11000 | |
| Deletion | Chr3 | 31114619 | 31114667 | 48 | |
| Deletion | Chr1 | 32945413 | 32945414 | 1 | LOC_Os01g57030 |
Insertions: 4
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 1946332 | 1946333 | 2 | |
| Insertion | Chr10 | 19526014 | 19526014 | 1 | |
| Insertion | Chr5 | 18832874 | 18832875 | 2 | |
| Insertion | Chr6 | 23530241 | 23530242 | 2 |
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr4 | 12053224 | 12520677 | LOC_Os04g22100 |
| Inversion | Chr2 | 20313474 | 20917314 | 2 |
Translocations: 8
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr6 | 8929372 | Chr3 | 6177641 | LOC_Os06g15740 |
| Translocation | Chr12 | 16035776 | Chr11 | 10835836 | |
| Translocation | Chr11 | 17697754 | Chr5 | 22429731 | |
| Translocation | Chr11 | 22020946 | Chr6 | 8627587 | LOC_Os11g37290 |
| Translocation | Chr5 | 24311296 | Chr4 | 14242312 | |
| Translocation | Chr8 | 27513544 | Chr3 | 2010397 | LOC_Os08g43500 |
| Translocation | Chr8 | 27514089 | Chr3 | 2007342 | |
| Translocation | Chr6 | 31242972 | Chr4 | 32540367 | 2 |
