Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN-283 [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN-283 Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 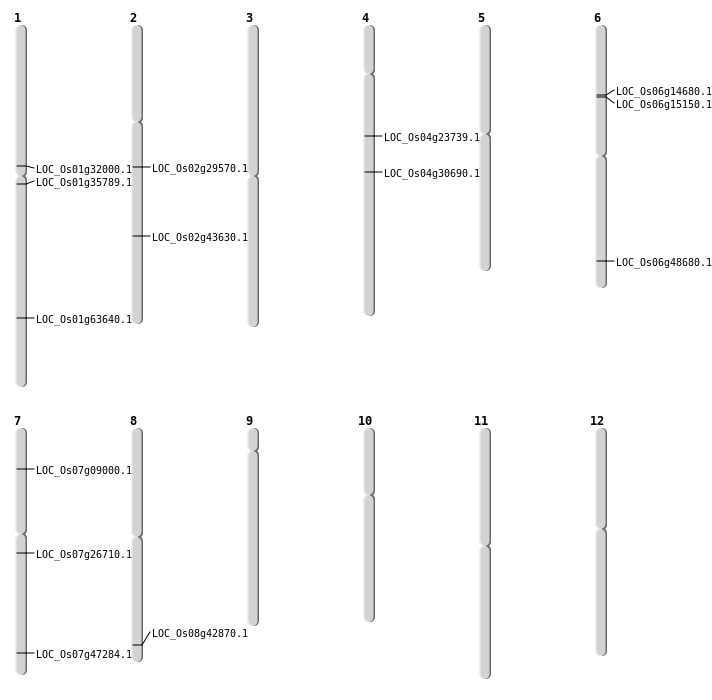
|
| Variant Information |
|---|
Single base substitutions: 40
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 25310024 | A-G | HOMO | INTERGENIC | |
| SBS | Chr1 | 32529276 | T-C | HET | INTERGENIC | |
| SBS | Chr1 | 36910778 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g63640 |
| SBS | Chr10 | 10322625 | G-C | HOMO | UTR_5_PRIME | |
| SBS | Chr10 | 16782870 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 21551806 | G-T | HOMO | INTERGENIC | |
| SBS | Chr10 | 58524 | C-A | HET | INTERGENIC | |
| SBS | Chr11 | 22326368 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 8226727 | C-A | HET | INTRON | |
| SBS | Chr12 | 6573556 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 14305086 | A-G | HET | INTERGENIC | |
| SBS | Chr2 | 14305087 | A-T | HET | INTERGENIC | |
| SBS | Chr2 | 15203667 | A-G | HET | INTERGENIC | |
| SBS | Chr2 | 18425232 | G-C | HET | INTERGENIC | |
| SBS | Chr2 | 19868282 | A-G | HET | UTR_3_PRIME | |
| SBS | Chr2 | 21309215 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 15212263 | A-G | HET | INTERGENIC | |
| SBS | Chr3 | 20440412 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 13103088 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 18341758 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g30690 |
| SBS | Chr4 | 19163657 | C-A | HET | INTERGENIC | |
| SBS | Chr4 | 24805511 | G-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 29007134 | G-A | HOMO | INTERGENIC | |
| SBS | Chr5 | 16840511 | G-T | HOMO | INTERGENIC | |
| SBS | Chr5 | 19589215 | C-T | HET | SPLICE_SITE_REGION | |
| SBS | Chr5 | 7587233 | G-A | HOMO | INTERGENIC | |
| SBS | Chr5 | 8519185 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr6 | 15204840 | A-C | HET | INTERGENIC | |
| SBS | Chr6 | 24313365 | T-C | HOMO | INTRON | |
| SBS | Chr6 | 30085261 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 8256876 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os06g14680 |
| SBS | Chr6 | 8587358 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os06g15150 |
| SBS | Chr7 | 17258358 | G-A | HET | INTRON | |
| SBS | Chr7 | 21964961 | A-G | HET | INTERGENIC | |
| SBS | Chr7 | 22999709 | T-A | HET | INTRON | |
| SBS | Chr7 | 28267326 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os07g47284 |
| SBS | Chr7 | 28267328 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os07g47284 |
| SBS | Chr7 | 4683870 | A-G | HOMO | NON_SYNONYMOUS_CODING | LOC_Os07g09000 |
| SBS | Chr8 | 13901771 | G-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr8 | 7525166 | T-A | HOMO | INTERGENIC |
Deletions: 18
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr10 | 1912043 | 1912044 | 1 | |
| Deletion | Chr6 | 3468380 | 3468399 | 19 | |
| Deletion | Chr4 | 5330440 | 5330449 | 9 | |
| Deletion | Chr12 | 6865152 | 6865156 | 4 | |
| Deletion | Chr4 | 11029158 | 11029161 | 3 | |
| Deletion | Chr8 | 11135826 | 11135837 | 11 | |
| Deletion | Chr3 | 14090980 | 14090986 | 6 | |
| Deletion | Chr2 | 15203594 | 15203613 | 19 | |
| Deletion | Chr7 | 15435839 | 15435872 | 33 | LOC_Os07g26710 |
| Deletion | Chr11 | 15612169 | 15612170 | 1 | |
| Deletion | Chr5 | 18613374 | 18613380 | 6 | |
| Deletion | Chr4 | 20618992 | 20618993 | 1 | |
| Deletion | Chr2 | 20655871 | 20655876 | 5 | |
| Deletion | Chr7 | 25509557 | 25509572 | 15 | |
| Deletion | Chr2 | 26196953 | 26196963 | 10 | |
| Deletion | Chr2 | 26329439 | 26329446 | 7 | LOC_Os02g43630 |
| Deletion | Chr8 | 27107151 | 27107450 | 299 | LOC_Os08g42870 |
| Deletion | Chr3 | 27539470 | 27539474 | 4 |
Insertions: 9
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 19824972 | 19824989 | 18 | LOC_Os01g35789 |
| Insertion | Chr10 | 9640700 | 9640700 | 1 | |
| Insertion | Chr11 | 26037933 | 26037934 | 2 | |
| Insertion | Chr11 | 4818876 | 4818879 | 4 | |
| Insertion | Chr5 | 6316576 | 6316581 | 6 | |
| Insertion | Chr6 | 24610240 | 24610241 | 2 | |
| Insertion | Chr6 | 29446738 | 29446738 | 1 | LOC_Os06g48680 |
| Insertion | Chr7 | 8787925 | 8787926 | 2 | |
| Insertion | Chr8 | 13065608 | 13065608 | 1 |
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr4 | 13577920 | 13578030 | LOC_Os04g23739 |
Translocations: 5
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr6 | 369047 | Chr1 | 42745749 | |
| Translocation | Chr2 | 17593638 | Chr1 | 17520919 | 2 |
| Translocation | Chr11 | 17927409 | Chr8 | 6186086 | |
| Translocation | Chr10 | 23044153 | Chr3 | 32469661 | |
| Translocation | Chr8 | 25167693 | Chr2 | 5772853 |
