Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN-287 [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN-287 Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 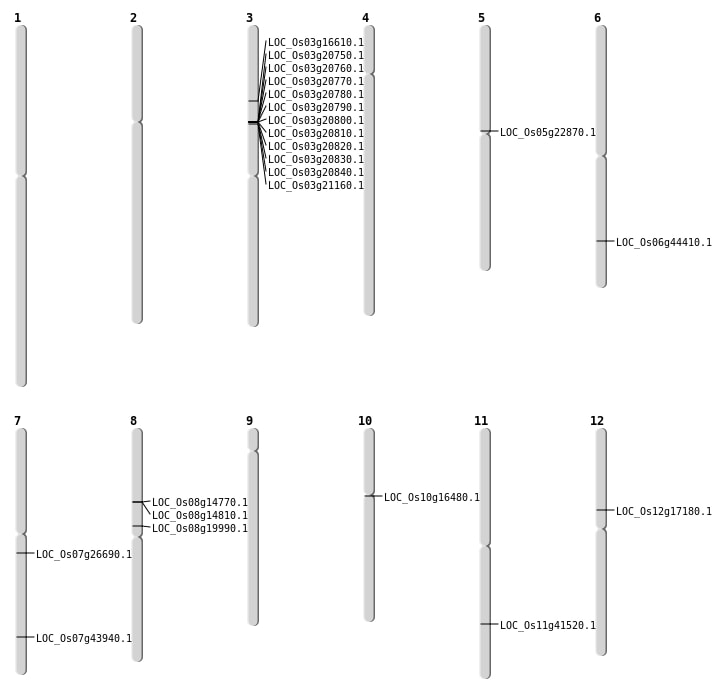
|
| Variant Information |
|---|
Single base substitutions: 27
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 2314789 | G-T | HET | INTERGENIC | |
| SBS | Chr1 | 26453043 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 42191959 | C-T | HET | SPLICE_SITE_REGION | |
| SBS | Chr10 | 22761855 | T-A | HET | INTERGENIC | |
| SBS | Chr10 | 3308685 | C-G | HOMO | INTERGENIC | |
| SBS | Chr12 | 14334991 | G-C | HET | INTRON | |
| SBS | Chr12 | 21720080 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 27342272 | A-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr2 | 18897204 | T-C | HET | INTERGENIC | |
| SBS | Chr4 | 10813531 | T-C | HET | INTERGENIC | |
| SBS | Chr4 | 15181030 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 20231394 | T-C | HET | INTERGENIC | |
| SBS | Chr4 | 20839093 | C-G | HET | INTERGENIC | |
| SBS | Chr4 | 35072679 | A-G | HET | SPLICE_SITE_REGION | |
| SBS | Chr5 | 10870428 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 11732895 | T-G | HET | INTERGENIC | |
| SBS | Chr6 | 26808461 | C-T | HET | STOP_GAINED | LOC_Os06g44410 |
| SBS | Chr6 | 31240342 | C-A | HET | INTERGENIC | |
| SBS | Chr7 | 15406621 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os07g26690 |
| SBS | Chr7 | 25442415 | C-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr7 | 9507318 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 17393358 | G-A | HET | INTERGENIC | |
| SBS | Chr8 | 19987271 | T-A | HOMO | INTRON | |
| SBS | Chr8 | 7186570 | G-T | HET | INTERGENIC | |
| SBS | Chr8 | 9614142 | C-T | HET | INTRON | |
| SBS | Chr9 | 14090338 | T-C | HET | INTERGENIC | |
| SBS | Chr9 | 8241336 | G-T | HET | INTERGENIC |
Deletions: 27
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr7 | 1516828 | 1516833 | 5 | |
| Deletion | Chr4 | 5208878 | 5208882 | 4 | |
| Deletion | Chr11 | 7479481 | 7479483 | 2 | |
| Deletion | Chr10 | 8220741 | 8220742 | 1 | LOC_Os10g16480 |
| Deletion | Chr3 | 9169307 | 9169331 | 24 | LOC_Os03g16610 |
| Deletion | Chr12 | 9839380 | 9839398 | 18 | LOC_Os12g17180 |
| Deletion | Chr9 | 10327343 | 10327352 | 9 | |
| Deletion | Chr1 | 10912205 | 10912210 | 5 | |
| Deletion | Chr3 | 11745001 | 11803000 | 58000 | 10 |
| Deletion | Chr8 | 11783791 | 11783797 | 6 | |
| Deletion | Chr8 | 11970048 | 11970078 | 30 | LOC_Os08g19990 |
| Deletion | Chr5 | 12990457 | 12990459 | 2 | LOC_Os05g22870 |
| Deletion | Chr7 | 14048357 | 14048375 | 18 | |
| Deletion | Chr2 | 15721186 | 15721187 | 1 | |
| Deletion | Chr2 | 17229679 | 17233864 | 4185 | |
| Deletion | Chr8 | 19995140 | 19995141 | 1 | |
| Deletion | Chr10 | 20244139 | 20244143 | 4 | |
| Deletion | Chr5 | 20968531 | 20968537 | 6 | |
| Deletion | Chr5 | 22318440 | 22318444 | 4 | |
| Deletion | Chr12 | 23849938 | 23849949 | 11 | |
| Deletion | Chr8 | 24517393 | 24517394 | 1 | |
| Deletion | Chr1 | 24648094 | 24648105 | 11 | |
| Deletion | Chr6 | 25158104 | 25158108 | 4 | |
| Deletion | Chr7 | 26260465 | 26261052 | 587 | LOC_Os07g43940 |
| Deletion | Chr5 | 26388197 | 26388211 | 14 | |
| Deletion | Chr6 | 28223311 | 28223316 | 5 | |
| Deletion | Chr1 | 41060027 | 41060040 | 13 |
Insertions: 10
Inversions: 4
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr3 | 11581189 | 11745311 | LOC_Os03g20750 |
| Inversion | Chr3 | 11802618 | 12071040 | LOC_Os03g21160 |
| Inversion | Chr2 | 18482608 | 26850073 | |
| Inversion | Chr11 | 24900190 | 24900496 | LOC_Os11g41520 |
Translocations: 10
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 3002781 | Chr10 | 13764672 | |
| Translocation | Chr3 | 3395198 | Chr2 | 18103409 | |
| Translocation | Chr5 | 6371170 | Chr4 | 16079080 | |
| Translocation | Chr8 | 8880197 | Chr2 | 17229760 | LOC_Os08g14770 |
| Translocation | Chr8 | 8880198 | Chr6 | 25131047 | LOC_Os08g14770 |
| Translocation | Chr8 | 8905206 | Chr6 | 25130870 | LOC_Os08g14810 |
| Translocation | Chr11 | 16229028 | Chr5 | 22549539 | |
| Translocation | Chr7 | 22081746 | Chr2 | 2430845 | |
| Translocation | Chr7 | 26260469 | Chr6 | 5342297 | LOC_Os07g43940 |
| Translocation | Chr7 | 26261029 | Chr6 | 5070946 | LOC_Os07g43940 |
Tandem duplications: 1
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Tandem Duplication | Chr3 | 12071036 | 12071036 | 0 | LOC_Os03g21160 |
