Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN-292 [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN-292 Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 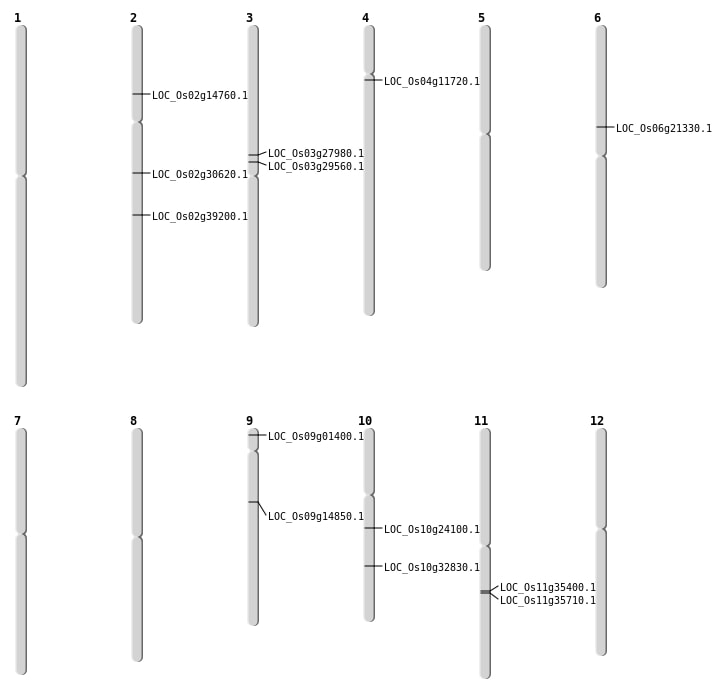
|
| Variant Information |
|---|
Single base substitutions: 30
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr10 | 12349371 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os10g24100 |
| SBS | Chr10 | 22263051 | T-A | HOMO | INTERGENIC | |
| SBS | Chr10 | 5799625 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 2534072 | A-T | HET | INTERGENIC | |
| SBS | Chr12 | 13168326 | T-C | HOMO | INTERGENIC | |
| SBS | Chr12 | 14579859 | C-A | HET | INTERGENIC | |
| SBS | Chr12 | 9803497 | G-C | HET | INTERGENIC | |
| SBS | Chr2 | 14825382 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 20731923 | C-G | HET | INTERGENIC | |
| SBS | Chr3 | 24045730 | A-G | HET | INTERGENIC | |
| SBS | Chr3 | 33127137 | C-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 361939 | T-C | HOMO | UTR_3_PRIME | |
| SBS | Chr3 | 361943 | T-A | HOMO | UTR_3_PRIME | |
| SBS | Chr4 | 1605980 | G-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr4 | 19377462 | A-C | HET | INTERGENIC | |
| SBS | Chr4 | 559327 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 6519464 | T-A | HET | INTERGENIC | |
| SBS | Chr6 | 12318935 | G-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os06g21330 |
| SBS | Chr6 | 31088614 | A-G | HOMO | INTRON | |
| SBS | Chr7 | 14324281 | G-A | HET | INTRON | |
| SBS | Chr7 | 18327144 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 18984209 | A-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 21873828 | G-A | HOMO | INTERGENIC | |
| SBS | Chr8 | 11782242 | C-T | HOMO | INTERGENIC | |
| SBS | Chr8 | 1364226 | A-C | HET | INTERGENIC | |
| SBS | Chr9 | 1672863 | G-C | HET | INTERGENIC | |
| SBS | Chr9 | 1672874 | T-A | HET | INTERGENIC | |
| SBS | Chr9 | 317516 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os09g01400 |
| SBS | Chr9 | 317518 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os09g01400 |
| SBS | Chr9 | 6685438 | T-C | HET | INTERGENIC |
Deletions: 17
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr2 | 1332520 | 1332526 | 6 | |
| Deletion | Chr12 | 4817624 | 4817631 | 7 | |
| Deletion | Chr8 | 6371527 | 6371533 | 6 | |
| Deletion | Chr4 | 6422247 | 6422309 | 62 | LOC_Os04g11720 |
| Deletion | Chr1 | 7156605 | 7156612 | 7 | |
| Deletion | Chr2 | 8165603 | 8167072 | 1469 | LOC_Os02g14760 |
| Deletion | Chr9 | 8860094 | 8860105 | 11 | LOC_Os09g14850 |
| Deletion | Chr6 | 10868357 | 10868415 | 58 | |
| Deletion | Chr10 | 16346031 | 16346033 | 2 | |
| Deletion | Chr3 | 16846233 | 16846240 | 7 | LOC_Os03g29560 |
| Deletion | Chr10 | 17188734 | 17188736 | 2 | LOC_Os10g32830 |
| Deletion | Chr7 | 17746317 | 17746357 | 40 | |
| Deletion | Chr2 | 18230172 | 18230173 | 1 | LOC_Os02g30620 |
| Deletion | Chr11 | 19604581 | 19606761 | 2180 | |
| Deletion | Chr2 | 23677792 | 23677793 | 1 | LOC_Os02g39200 |
| Deletion | Chr6 | 25593139 | 25593158 | 19 | |
| Deletion | Chr3 | 29041791 | 29041796 | 5 |
Insertions: 1
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr2 | 3959736 | 3959736 | 1 |
Inversions: 4
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr12 | 15009328 | 15089625 | |
| Inversion | Chr3 | 16072332 | 16357308 | LOC_Os03g27980 |
| Inversion | Chr11 | 20747972 | 20952082 | 2 |
| Inversion | Chr11 | 20748003 | 20952088 | 2 |
Translocations: 1
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr7 | 14276134 | Chr2 | 8165615 | LOC_Os02g14760 |
