Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN-315 [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN-315 Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 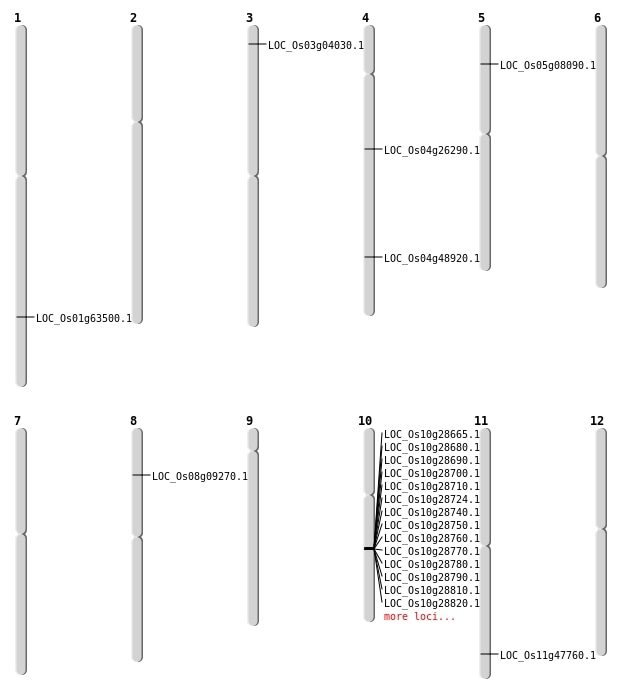
|
| Variant Information |
|---|
Single base substitutions: 30
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 31983844 | G-C | HOMO | INTERGENIC | |
| SBS | Chr1 | 36808585 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os01g63500 |
| SBS | Chr10 | 13337630 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 13337638 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 15363767 | G-T | HOMO | STOP_GAINED | LOC_Os10g29560 |
| SBS | Chr10 | 15363768 | A-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os10g29560 |
| SBS | Chr10 | 17686108 | C-G | HET | INTERGENIC | |
| SBS | Chr10 | 22213960 | C-T | HET | INTRON | |
| SBS | Chr11 | 28807153 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os11g47760 |
| SBS | Chr12 | 11919127 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 11385050 | C-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 32559293 | C-T | HET | INTRON | |
| SBS | Chr3 | 12934779 | G-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 1838424 | C-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os03g04030 |
| SBS | Chr4 | 15325223 | C-A | HOMO | STOP_GAINED | LOC_Os04g26290 |
| SBS | Chr4 | 1605980 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr4 | 19045630 | C-T | HOMO | INTRON | |
| SBS | Chr4 | 25149271 | A-G | HOMO | INTRON | |
| SBS | Chr5 | 20369899 | T-A | HOMO | INTERGENIC | |
| SBS | Chr5 | 4996872 | T-A | HET | INTERGENIC | |
| SBS | Chr5 | 8147325 | T-A | HET | INTERGENIC | |
| SBS | Chr6 | 20211010 | G-A | HOMO | INTERGENIC | |
| SBS | Chr6 | 27589237 | G-A | HOMO | INTRON | |
| SBS | Chr6 | 28349617 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os06g46670 |
| SBS | Chr6 | 28430656 | A-G | HOMO | INTERGENIC | |
| SBS | Chr7 | 23427933 | G-A | HOMO | INTRON | |
| SBS | Chr7 | 5973416 | T-G | HET | INTRON | |
| SBS | Chr8 | 8046584 | A-T | HET | INTERGENIC | |
| SBS | Chr9 | 21615299 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 5171851 | G-T | HOMO | INTERGENIC |
Deletions: 8
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr8 | 5385907 | 5385909 | 2 | LOC_Os08g09270 |
| Deletion | Chr10 | 14950001 | 15052000 | 102000 | 20 |
| Deletion | Chr10 | 15055001 | 15953000 | 898000 | 158 |
| Deletion | Chr10 | 15970001 | 16437000 | 467000 | 54 |
| Deletion | Chr9 | 17870669 | 17870676 | 7 | |
| Deletion | Chr10 | 20660001 | 20724000 | 64000 | 7 |
| Deletion | Chr8 | 20862594 | 20862597 | 3 | |
| Deletion | Chr4 | 29177228 | 29177234 | 6 | LOC_Os04g48920 |
Insertions: 2
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr10 | 7248748 | 7248748 | 1 | |
| Insertion | Chr9 | 21908464 | 21908464 | 1 |
Inversions: 4
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr5 | 4157988 | 5113967 | |
| Inversion | Chr5 | 4158030 | 4405185 | LOC_Os05g08090 |
| Inversion | Chr2 | 14448429 | 14459672 | |
| Inversion | Chr2 | 14448445 | 14459686 |
No Translocation
