Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN-335 [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN-335 Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 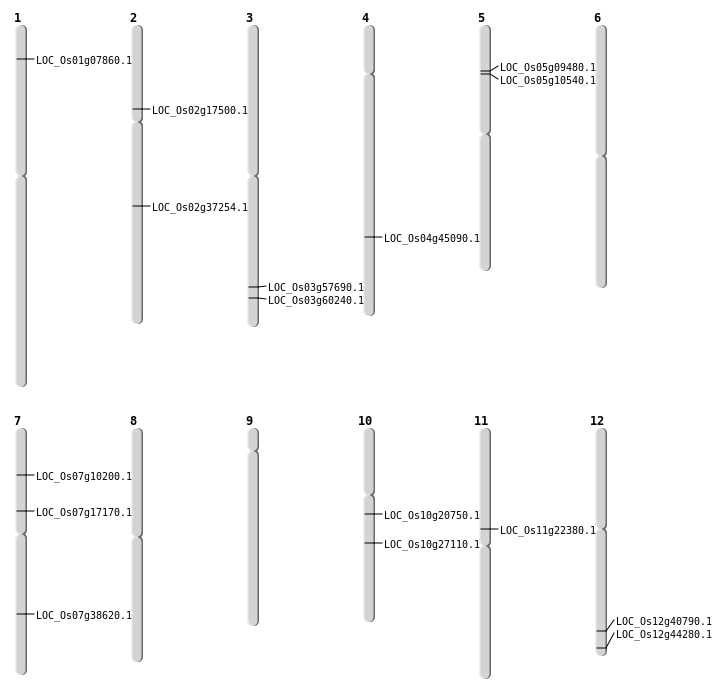
|
| Variant Information |
|---|
Single base substitutions: 41
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 13878753 | C-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 38041961 | C-T | HET | INTRON | |
| SBS | Chr1 | 5170020 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 7568839 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 10517083 | G-C | HOMO | NON_SYNONYMOUS_CODING | LOC_Os10g20750 |
| SBS | Chr10 | 13640841 | G-T | HOMO | INTERGENIC | |
| SBS | Chr10 | 15995155 | A-G | HOMO | INTERGENIC | |
| SBS | Chr10 | 1765985 | T-A | HET | INTERGENIC | |
| SBS | Chr11 | 12836226 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os11g22380 |
| SBS | Chr11 | 13728548 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 15345876 | A-T | HET | INTERGENIC | |
| SBS | Chr11 | 15577217 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 12528677 | T-C | HET | INTERGENIC | |
| SBS | Chr12 | 25050252 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 25250301 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os12g40790 |
| SBS | Chr12 | 27446979 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os12g44280 |
| SBS | Chr12 | 5480418 | A-T | HET | INTERGENIC | |
| SBS | Chr2 | 10416176 | G-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 15878069 | A-G | HOMO | INTERGENIC | |
| SBS | Chr2 | 21599967 | A-T | HET | INTRON | |
| SBS | Chr3 | 14799387 | T-A | HET | INTERGENIC | |
| SBS | Chr3 | 35538309 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 763340 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 16827767 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 23737753 | G-T | HET | INTRON | |
| SBS | Chr4 | 25068562 | A-G | HET | INTRON | |
| SBS | Chr5 | 10725039 | C-T | HET | INTRON | |
| SBS | Chr5 | 5728893 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os05g10540 |
| SBS | Chr6 | 10146362 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 7410395 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 907564 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 18811758 | A-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 18811760 | A-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 20922143 | C-A | HET | INTERGENIC | |
| SBS | Chr7 | 23199582 | G-C | HET | INTERGENIC | |
| SBS | Chr7 | 25446474 | A-T | HET | INTERGENIC | |
| SBS | Chr7 | 5485502 | C-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os07g10200 |
| SBS | Chr7 | 6368225 | C-G | HOMO | INTERGENIC | |
| SBS | Chr9 | 20708310 | G-T | HET | INTERGENIC | |
| SBS | Chr9 | 20708311 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 22921082 | T-C | HET | INTRON |
Deletions: 22
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr3 | 3461458 | 3461851 | 393 | |
| Deletion | Chr12 | 3799431 | 3799432 | 1 | |
| Deletion | Chr5 | 5331316 | 5331320 | 4 | LOC_Os05g09480 |
| Deletion | Chr8 | 9170029 | 9170034 | 5 | |
| Deletion | Chr11 | 9797532 | 9797549 | 17 | |
| Deletion | Chr2 | 10074660 | 10074661 | 1 | LOC_Os02g17500 |
| Deletion | Chr10 | 14289096 | 14289103 | 7 | LOC_Os10g27110 |
| Deletion | Chr8 | 15457663 | 15457677 | 14 | |
| Deletion | Chr11 | 15536965 | 15537004 | 39 | |
| Deletion | Chr1 | 16910236 | 16910243 | 7 | |
| Deletion | Chr1 | 17822712 | 17822713 | 1 | |
| Deletion | Chr1 | 20403062 | 20403080 | 18 | |
| Deletion | Chr2 | 22514839 | 22514842 | 3 | LOC_Os02g37254 |
| Deletion | Chr11 | 24241474 | 24241475 | 1 | |
| Deletion | Chr7 | 26360278 | 26360784 | 506 | |
| Deletion | Chr4 | 26675079 | 26675080 | 1 | LOC_Os04g45090 |
| Deletion | Chr7 | 28875945 | 28875946 | 1 | |
| Deletion | Chr1 | 30905367 | 30905369 | 2 | |
| Deletion | Chr3 | 31477974 | 31477993 | 19 | |
| Deletion | Chr3 | 32020800 | 32020814 | 14 | |
| Deletion | Chr3 | 34258234 | 34259423 | 1189 | LOC_Os03g60240 |
| Deletion | Chr3 | 35319588 | 35319589 | 1 |
Insertions: 3
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 39804019 | 39804020 | 2 | |
| Insertion | Chr3 | 16708020 | 16708021 | 2 | |
| Insertion | Chr7 | 1455532 | 1455543 | 12 |
Inversions: 7
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr1 | 3783901 | 5169919 | LOC_Os01g07860 |
| Inversion | Chr7 | 7450857 | 10097744 | |
| Inversion | Chr7 | 7451026 | 10098265 | LOC_Os07g17170 |
| Inversion | Chr7 | 10097747 | 23199001 | LOC_Os07g38620 |
| Inversion | Chr7 | 10098263 | 23198996 | 2 |
| Inversion | Chr3 | 26228155 | 34259386 | LOC_Os03g60240 |
| Inversion | Chr3 | 32654658 | 32883880 | LOC_Os03g57690 |
Translocations: 1
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 9725498 | Chr4 | 10817551 |
