Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN-408 [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN-408 Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 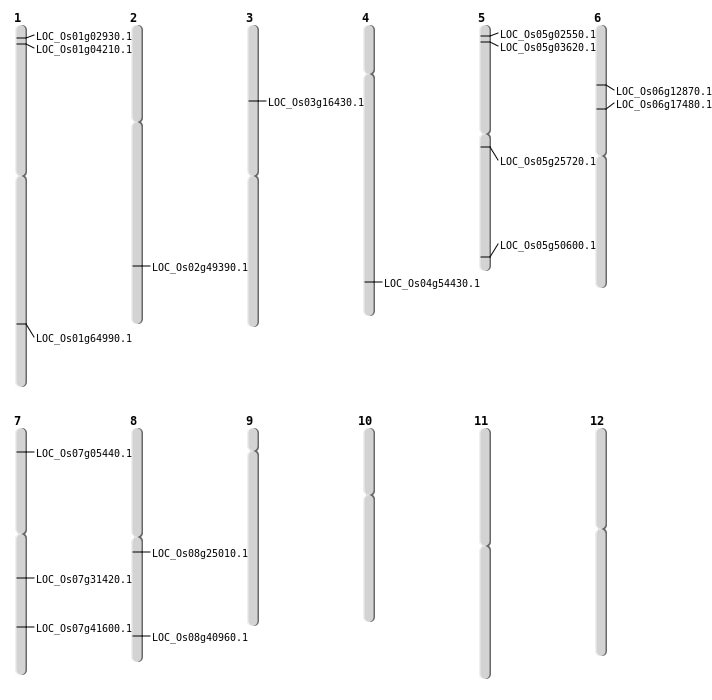
|
| Variant Information |
|---|
Single base substitutions: 31
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 11422319 | C-A | HET | INTERGENIC | |
| SBS | Chr1 | 15963419 | C-A | HET | INTERGENIC | |
| SBS | Chr1 | 1857998 | A-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os01g04210 |
| SBS | Chr1 | 2015679 | C-T | HOMO | INTRON | |
| SBS | Chr1 | 2590346 | C-T | HOMO | INTRON | |
| SBS | Chr1 | 36824979 | A-T | HET | INTRON | |
| SBS | Chr10 | 12001228 | T-A | HOMO | INTERGENIC | |
| SBS | Chr12 | 13896077 | T-C | HOMO | INTRON | |
| SBS | Chr12 | 25417515 | G-C | HOMO | INTRON | |
| SBS | Chr12 | 8063608 | G-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 1521477 | C-A | HET | INTRON | |
| SBS | Chr2 | 28149650 | A-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr2 | 30178052 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os02g49390 |
| SBS | Chr2 | 390926 | A-G | HET | INTERGENIC | |
| SBS | Chr3 | 33577503 | A-T | HET | INTERGENIC | |
| SBS | Chr3 | 9065515 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os03g16430 |
| SBS | Chr3 | 9065516 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os03g16430 |
| SBS | Chr4 | 12497324 | G-T | HET | INTERGENIC | |
| SBS | Chr4 | 19045427 | T-A | HOMO | INTRON | |
| SBS | Chr5 | 14963875 | T-A | HET | STOP_LOST | LOC_Os05g25720 |
| SBS | Chr5 | 18226647 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 29004962 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os05g50600 |
| SBS | Chr6 | 29767541 | A-G | HOMO | INTERGENIC | |
| SBS | Chr6 | 29771109 | A-G | HOMO | INTRON | |
| SBS | Chr6 | 29771111 | A-T | HOMO | INTRON | |
| SBS | Chr7 | 15942843 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 2185581 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 2530624 | G-T | HET | INTERGENIC | |
| SBS | Chr8 | 19895117 | G-T | HET | INTERGENIC | |
| SBS | Chr8 | 19895118 | T-G | HET | INTERGENIC | |
| SBS | Chr9 | 6751112 | C-G | HET | INTERGENIC |
Deletions: 30
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr6 | 116856 | 116857 | 1 | |
| Deletion | Chr11 | 129608 | 129612 | 4 | |
| Deletion | Chr10 | 654079 | 654083 | 4 | |
| Deletion | Chr5 | 885567 | 885576 | 9 | LOC_Os05g02550 |
| Deletion | Chr1 | 1081427 | 1081431 | 4 | LOC_Os01g02930 |
| Deletion | Chr7 | 2506477 | 2506485 | 8 | LOC_Os07g05440 |
| Deletion | Chr4 | 4258346 | 4258365 | 19 | |
| Deletion | Chr12 | 4835806 | 4835807 | 1 | |
| Deletion | Chr5 | 6688065 | 6688066 | 1 | |
| Deletion | Chr6 | 7044498 | 7044507 | 9 | LOC_Os06g12870 |
| Deletion | Chr4 | 7877144 | 7877145 | 1 | |
| Deletion | Chr9 | 10079654 | 10079809 | 155 | |
| Deletion | Chr9 | 10079655 | 10079810 | 155 | |
| Deletion | Chr6 | 10138541 | 10138817 | 276 | LOC_Os06g17480 |
| Deletion | Chr8 | 12384201 | 12384202 | 1 | |
| Deletion | Chr2 | 14102905 | 14102907 | 2 | |
| Deletion | Chr4 | 14651648 | 14651655 | 7 | |
| Deletion | Chr8 | 15166455 | 15166467 | 12 | LOC_Os08g25010 |
| Deletion | Chr7 | 18612145 | 18612186 | 41 | LOC_Os07g31420 |
| Deletion | Chr11 | 19056033 | 19056034 | 1 | |
| Deletion | Chr5 | 21346654 | 21346657 | 3 | |
| Deletion | Chr3 | 24467377 | 24467378 | 1 | |
| Deletion | Chr8 | 25917388 | 25917409 | 21 | LOC_Os08g40960 |
| Deletion | Chr12 | 26189110 | 26189118 | 8 | |
| Deletion | Chr12 | 27163347 | 27163354 | 7 | |
| Deletion | Chr4 | 27534991 | 27534992 | 1 | |
| Deletion | Chr3 | 28591493 | 28591497 | 4 | |
| Deletion | Chr6 | 29041017 | 29041046 | 29 | |
| Deletion | Chr4 | 32384562 | 32384570 | 8 | LOC_Os04g54430 |
| Deletion | Chr1 | 33780725 | 33780728 | 3 |
Insertions: 37
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 11584081 | 11584088 | 8 | |
| Insertion | Chr1 | 23494213 | 23494214 | 2 | |
| Insertion | Chr1 | 23745436 | 23745439 | 4 | |
| Insertion | Chr1 | 3341686 | 3341687 | 2 | |
| Insertion | Chr1 | 6154084 | 6154087 | 4 | |
| Insertion | Chr10 | 14175586 | 14175587 | 2 | |
| Insertion | Chr10 | 3986710 | 3986711 | 2 | |
| Insertion | Chr11 | 16919102 | 16919103 | 2 | |
| Insertion | Chr11 | 23208034 | 23208036 | 3 | |
| Insertion | Chr11 | 23454869 | 23454869 | 1 | |
| Insertion | Chr11 | 28281687 | 28281690 | 4 | |
| Insertion | Chr11 | 4818876 | 4818879 | 4 | |
| Insertion | Chr12 | 9349145 | 9349146 | 2 | |
| Insertion | Chr12 | 9977598 | 9977608 | 11 | |
| Insertion | Chr2 | 16516655 | 16516656 | 2 | |
| Insertion | Chr2 | 16607553 | 16607554 | 2 | |
| Insertion | Chr2 | 18534196 | 18534197 | 2 | |
| Insertion | Chr2 | 23138618 | 23138618 | 1 | |
| Insertion | Chr2 | 26650879 | 26650880 | 2 | |
| Insertion | Chr3 | 16732248 | 16732249 | 2 | |
| Insertion | Chr3 | 25283995 | 25283996 | 2 | |
| Insertion | Chr3 | 7620255 | 7620256 | 2 | |
| Insertion | Chr4 | 17630012 | 17630014 | 3 | |
| Insertion | Chr4 | 34730832 | 34730835 | 4 | |
| Insertion | Chr5 | 22078777 | 22078782 | 6 | |
| Insertion | Chr5 | 6402287 | 6402290 | 4 | |
| Insertion | Chr5 | 7120260 | 7120261 | 2 | |
| Insertion | Chr6 | 25066086 | 25066086 | 1 | |
| Insertion | Chr7 | 20230947 | 20230947 | 1 | |
| Insertion | Chr7 | 24934517 | 24934519 | 3 | LOC_Os07g41600 |
| Insertion | Chr7 | 9078140 | 9078140 | 1 | |
| Insertion | Chr7 | 9965619 | 9965619 | 1 | |
| Insertion | Chr8 | 18164358 | 18164359 | 2 | |
| Insertion | Chr8 | 26353245 | 26353250 | 6 | |
| Insertion | Chr8 | 3638362 | 3638363 | 2 | |
| Insertion | Chr8 | 8336123 | 8336124 | 2 | |
| Insertion | Chr9 | 18750952 | 18750953 | 2 |
Inversions: 3
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr5 | 1547562 | 1686493 | LOC_Os05g03620 |
| Inversion | Chr7 | 13586659 | 14559752 | |
| Inversion | Chr1 | 37519374 | 37725591 | LOC_Os01g64990 |
No Translocation
