Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN-409 [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN-409 Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 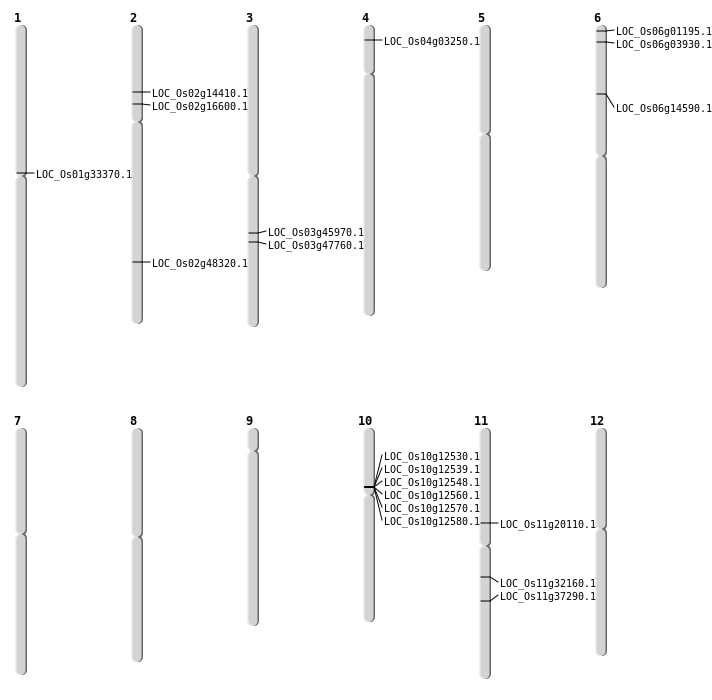
|
| Variant Information |
|---|
Single base substitutions: 39
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 10943780 | G-T | HET | INTERGENIC | |
| SBS | Chr1 | 13780507 | T-C | HET | INTERGENIC | |
| SBS | Chr1 | 13780513 | A-T | HET | INTERGENIC | |
| SBS | Chr1 | 15486396 | C-A | HET | INTRON | |
| SBS | Chr1 | 17107056 | A-T | HET | INTRON | |
| SBS | Chr1 | 18376534 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g33370 |
| SBS | Chr1 | 31098056 | T-C | HOMO | INTERGENIC | |
| SBS | Chr11 | 22019466 | G-A | HOMO | STOP_GAINED | LOC_Os11g37290 |
| SBS | Chr11 | 8533468 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 12729460 | G-T | HET | INTERGENIC | |
| SBS | Chr2 | 3468886 | A-G | HET | UTR_3_PRIME | |
| SBS | Chr2 | 8344704 | A-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 9492216 | G-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os02g16600 |
| SBS | Chr2 | 9492218 | C-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr3 | 18817260 | A-G | HOMO | INTERGENIC | |
| SBS | Chr3 | 27105821 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g47760 |
| SBS | Chr3 | 28233086 | T-C | HET | INTRON | |
| SBS | Chr3 | 30509984 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 21215831 | G-A | HOMO | UTR_3_PRIME | |
| SBS | Chr4 | 2286021 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 34338379 | A-G | HET | INTERGENIC | |
| SBS | Chr4 | 6184693 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr5 | 10492316 | C-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr5 | 12696242 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 12696243 | C-A | HET | INTERGENIC | |
| SBS | Chr5 | 27508126 | G-A | HET | INTRON | |
| SBS | Chr5 | 27508128 | T-A | HET | INTRON | |
| SBS | Chr5 | 9662277 | C-T | HET | INTRON | |
| SBS | Chr6 | 10661390 | A-T | HOMO | INTERGENIC | |
| SBS | Chr6 | 140931 | G-C | HET | INTERGENIC | |
| SBS | Chr6 | 8212931 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os06g14590 |
| SBS | Chr7 | 7892430 | G-A | HET | INTERGENIC | |
| SBS | Chr8 | 11592499 | C-A | HET | INTERGENIC | |
| SBS | Chr8 | 15588880 | C-A | HET | INTERGENIC | |
| SBS | Chr8 | 5194750 | G-A | HOMO | INTERGENIC | |
| SBS | Chr9 | 20644583 | T-A | HET | INTERGENIC | |
| SBS | Chr9 | 2180782 | G-T | HET | INTERGENIC | |
| SBS | Chr9 | 2180783 | A-T | HET | INTERGENIC | |
| SBS | Chr9 | 261368 | G-T | HET | INTERGENIC |
Deletions: 24
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr3 | 460218 | 460235 | 17 | |
| Deletion | Chr1 | 967685 | 967688 | 3 | |
| Deletion | Chr3 | 1252392 | 1252412 | 20 | |
| Deletion | Chr12 | 2677932 | 2677939 | 7 | |
| Deletion | Chr2 | 4168832 | 4168841 | 9 | |
| Deletion | Chr10 | 6966001 | 7008000 | 42000 | 6 |
| Deletion | Chr6 | 7279455 | 7279456 | 1 | |
| Deletion | Chr8 | 7608567 | 7608568 | 1 | |
| Deletion | Chr8 | 8065325 | 8065333 | 8 | |
| Deletion | Chr12 | 8193894 | 8193904 | 10 | |
| Deletion | Chr6 | 10564006 | 10564009 | 3 | |
| Deletion | Chr11 | 11618466 | 11618475 | 9 | LOC_Os11g20110 |
| Deletion | Chr12 | 16357617 | 16357722 | 105 | |
| Deletion | Chr11 | 19282698 | 19282699 | 1 | |
| Deletion | Chr3 | 21772371 | 21772377 | 6 | |
| Deletion | Chr12 | 23057849 | 23057850 | 1 | |
| Deletion | Chr8 | 24798578 | 24798579 | 1 | |
| Deletion | Chr3 | 26444842 | 26444852 | 10 | |
| Deletion | Chr6 | 28017902 | 28017911 | 9 | |
| Deletion | Chr3 | 29388538 | 29388588 | 50 | |
| Deletion | Chr2 | 29574143 | 29574149 | 6 | LOC_Os02g48320 |
| Deletion | Chr1 | 31098463 | 31098464 | 1 | |
| Deletion | Chr1 | 33408643 | 33408668 | 25 | |
| Deletion | Chr3 | 35295838 | 35295843 | 5 |
No Insertion
Inversions: 7
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr6 | 141356 | 1582950 | LOC_Os06g03930 |
| Inversion | Chr6 | 141374 | 1582989 | 2 |
| Inversion | Chr4 | 14606774 | 17125241 | |
| Inversion | Chr11 | 18792781 | 18987906 | LOC_Os11g32160 |
| Inversion | Chr12 | 23204828 | 23975605 | |
| Inversion | Chr3 | 25973924 | 25978876 | LOC_Os03g45970 |
| Inversion | Chr3 | 25973957 | 25978901 | LOC_Os03g45970 |
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr4 | 1384924 | Chr2 | 7913690 | 2 |
| Translocation | Chr9 | 1446404 | Chr4 | 8961751 |
