Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN-70 [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN-70 Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 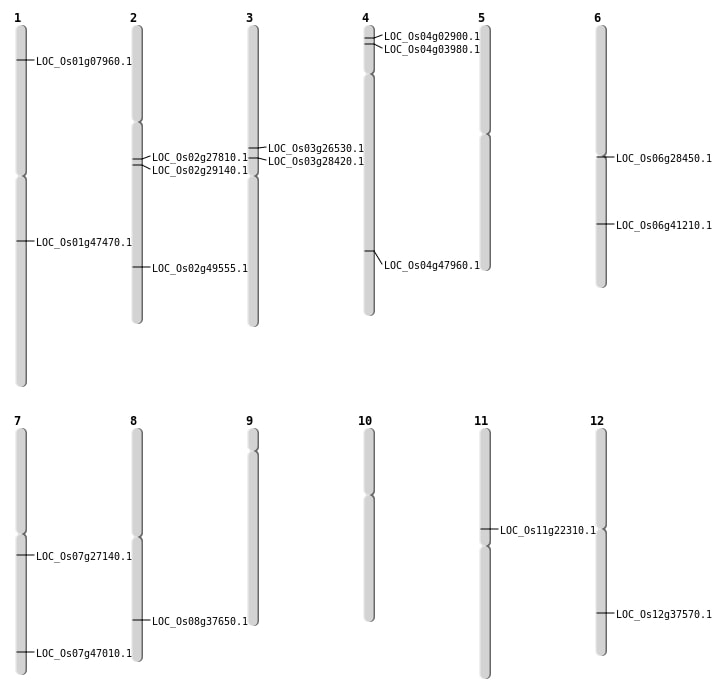
|
| Variant Information |
|---|
Single base substitutions: 31
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 12682467 | T-G | HET | INTRON | |
| SBS | Chr1 | 27180197 | A-T | HOMO | INTRON | |
| SBS | Chr10 | 12206886 | T-C | HET | INTERGENIC | |
| SBS | Chr10 | 20510952 | C-G | HOMO | INTERGENIC | |
| SBS | Chr11 | 12788717 | G-A | HOMO | STOP_GAINED | LOC_Os11g22310 |
| SBS | Chr11 | 17578997 | A-C | HET | INTRON | |
| SBS | Chr11 | 24908329 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 16862391 | A-T | HET | INTERGENIC | |
| SBS | Chr12 | 23063558 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os12g37570 |
| SBS | Chr12 | 27207326 | C-A | HET | INTERGENIC | |
| SBS | Chr2 | 29495590 | G-C | HOMO | INTERGENIC | |
| SBS | Chr3 | 10789482 | A-C | HET | INTERGENIC | |
| SBS | Chr3 | 11534106 | T-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 11534114 | T-C | HOMO | INTERGENIC | |
| SBS | Chr3 | 16835795 | C-A | HET | INTERGENIC | |
| SBS | Chr3 | 25780585 | T-C | HOMO | INTERGENIC | |
| SBS | Chr3 | 27054328 | T-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 13625697 | G-T | HET | INTERGENIC | |
| SBS | Chr4 | 13625698 | G-T | HET | INTERGENIC | |
| SBS | Chr4 | 15280456 | C-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr4 | 1815338 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g03980 |
| SBS | Chr4 | 19950274 | G-T | HOMO | INTERGENIC | |
| SBS | Chr4 | 21163227 | C-T | HOMO | INTERGENIC | |
| SBS | Chr4 | 26258120 | C-A | HOMO | UTR_3_PRIME | |
| SBS | Chr5 | 8767451 | C-T | HET | INTRON | |
| SBS | Chr6 | 13525600 | C-T | HET | INTRON | |
| SBS | Chr6 | 24670146 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os06g41210 |
| SBS | Chr6 | 27894913 | A-G | HET | INTERGENIC | |
| SBS | Chr7 | 12984225 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 22951283 | C-T | HOMO | INTERGENIC | |
| SBS | Chr9 | 21046801 | G-A | HOMO | UTR_5_PRIME |
Deletions: 33
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr6 | 1118471 | 1118472 | 1 | |
| Deletion | Chr4 | 1136577 | 1136585 | 8 | LOC_Os04g02900 |
| Deletion | Chr1 | 2319385 | 2319402 | 17 | |
| Deletion | Chr1 | 3849226 | 3849236 | 10 | LOC_Os01g07960 |
| Deletion | Chr9 | 4224416 | 4224423 | 7 | |
| Deletion | Chr12 | 7397732 | 7397736 | 4 | |
| Deletion | Chr9 | 9735385 | 9735393 | 8 | |
| Deletion | Chr11 | 10586294 | 10586297 | 3 | |
| Deletion | Chr3 | 11486232 | 11486245 | 13 | |
| Deletion | Chr11 | 11544346 | 11544374 | 28 | |
| Deletion | Chr4 | 14065772 | 14065777 | 5 | |
| Deletion | Chr3 | 15150684 | 15150693 | 9 | LOC_Os03g26530 |
| Deletion | Chr6 | 16183186 | 16183193 | 7 | LOC_Os06g28450 |
| Deletion | Chr3 | 16377301 | 16377308 | 7 | LOC_Os03g28420 |
| Deletion | Chr12 | 16852104 | 16852106 | 2 | |
| Deletion | Chr4 | 16944201 | 16944202 | 1 | |
| Deletion | Chr2 | 17262407 | 17262408 | 1 | LOC_Os02g29140 |
| Deletion | Chr11 | 17570807 | 17570808 | 1 | |
| Deletion | Chr10 | 18352166 | 18352167 | 1 | |
| Deletion | Chr4 | 18392004 | 18392080 | 76 | |
| Deletion | Chr1 | 19189943 | 19189951 | 8 | |
| Deletion | Chr1 | 21369948 | 21369952 | 4 | |
| Deletion | Chr4 | 22028933 | 22028967 | 34 | |
| Deletion | Chr8 | 22248809 | 22248820 | 11 | |
| Deletion | Chr7 | 22747635 | 22747637 | 2 | |
| Deletion | Chr8 | 23850651 | 23850657 | 6 | LOC_Os08g37650 |
| Deletion | Chr3 | 24195937 | 24196100 | 163 | |
| Deletion | Chr5 | 24453442 | 24453446 | 4 | |
| Deletion | Chr8 | 27623137 | 27623143 | 6 | |
| Deletion | Chr4 | 28087366 | 28087367 | 1 | |
| Deletion | Chr7 | 28091298 | 28091319 | 21 | LOC_Os07g47010 |
| Deletion | Chr4 | 28519420 | 28519722 | 302 | LOC_Os04g47960 |
| Deletion | Chr3 | 30145856 | 30145859 | 3 |
Insertions: 33
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 27134019 | 27134020 | 2 | LOC_Os01g47470 |
| Insertion | Chr1 | 27891734 | 27891735 | 2 | |
| Insertion | Chr1 | 39804019 | 39804020 | 2 | |
| Insertion | Chr1 | 43242276 | 43242281 | 6 | |
| Insertion | Chr1 | 8956020 | 8956021 | 2 | |
| Insertion | Chr10 | 18961710 | 18961711 | 2 | |
| Insertion | Chr10 | 5961878 | 5961879 | 2 | |
| Insertion | Chr11 | 7535667 | 7535674 | 8 | |
| Insertion | Chr12 | 18350022 | 18350023 | 2 | |
| Insertion | Chr12 | 21795985 | 21795986 | 2 | |
| Insertion | Chr2 | 16468454 | 16468457 | 4 | LOC_Os02g27810 |
| Insertion | Chr2 | 25361513 | 25361514 | 2 | |
| Insertion | Chr2 | 28851153 | 28851159 | 7 | |
| Insertion | Chr2 | 29249980 | 29249980 | 1 | |
| Insertion | Chr2 | 30290909 | 30290911 | 3 | LOC_Os02g49555 |
| Insertion | Chr2 | 6394878 | 6394879 | 2 | |
| Insertion | Chr3 | 4143849 | 4143864 | 16 | |
| Insertion | Chr4 | 2305618 | 2305618 | 1 | |
| Insertion | Chr4 | 31914425 | 31914428 | 4 | |
| Insertion | Chr5 | 6402287 | 6402290 | 4 | |
| Insertion | Chr7 | 12108498 | 12108500 | 3 | |
| Insertion | Chr7 | 15737652 | 15737652 | 1 | LOC_Os07g27140 |
| Insertion | Chr7 | 25272927 | 25272929 | 3 | |
| Insertion | Chr7 | 29332646 | 29332647 | 2 | |
| Insertion | Chr7 | 5316619 | 5316619 | 1 | |
| Insertion | Chr8 | 1209775 | 1209784 | 10 | |
| Insertion | Chr8 | 24023471 | 24023473 | 3 | |
| Insertion | Chr8 | 26815767 | 26815768 | 2 | |
| Insertion | Chr9 | 10486654 | 10486655 | 2 | |
| Insertion | Chr9 | 18154253 | 18154258 | 6 | |
| Insertion | Chr9 | 18174528 | 18174533 | 6 | |
| Insertion | Chr9 | 20022822 | 20022822 | 1 | |
| Insertion | Chr9 | 9197671 | 9197672 | 2 |
No Inversion
No Translocation
