Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN-71 [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN-71 Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 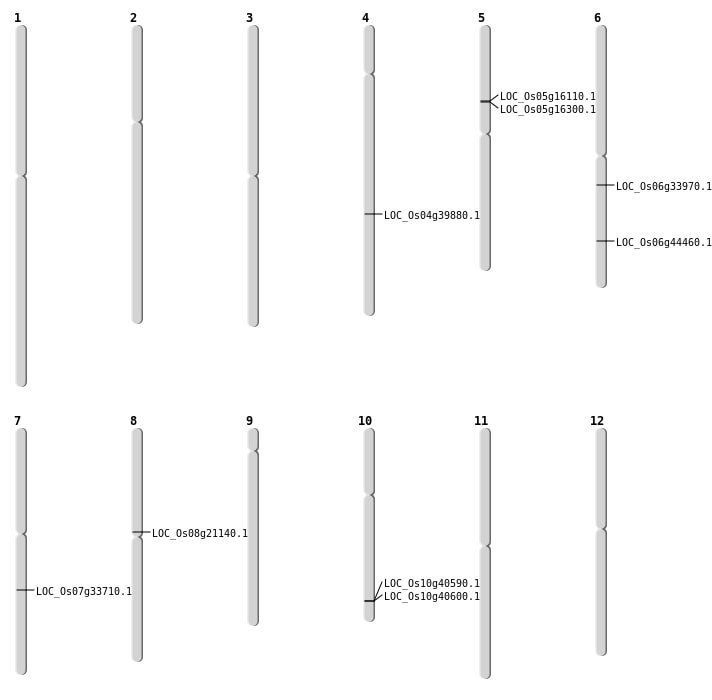
|
| Variant Information |
|---|
Single base substitutions: 38
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 24106645 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 41136975 | T-A | HET | INTERGENIC | |
| SBS | Chr1 | 7247453 | T-A | HET | INTERGENIC | |
| SBS | Chr10 | 14167524 | T-C | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr10 | 14174605 | A-G | HOMO | INTERGENIC | |
| SBS | Chr10 | 1702930 | A-T | HET | UTR_5_PRIME | |
| SBS | Chr10 | 22540191 | T-A | HET | INTERGENIC | |
| SBS | Chr11 | 15641551 | C-A | HOMO | INTRON | |
| SBS | Chr11 | 778173 | C-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 16821976 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 151486 | G-C | HET | INTERGENIC | |
| SBS | Chr3 | 19099719 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 30215260 | T-C | HOMO | INTERGENIC | |
| SBS | Chr3 | 8487859 | C-T | HET | UTR_5_PRIME | |
| SBS | Chr3 | 8487860 | C-T | HET | UTR_5_PRIME | |
| SBS | Chr4 | 16942573 | T-C | HOMO | INTERGENIC | |
| SBS | Chr4 | 19656896 | A-T | HOMO | INTERGENIC | |
| SBS | Chr4 | 19656897 | C-T | HOMO | INTERGENIC | |
| SBS | Chr4 | 24825895 | C-T | HOMO | INTRON | |
| SBS | Chr5 | 12122345 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 21605272 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 24239037 | T-A | HET | INTERGENIC | |
| SBS | Chr6 | 16355098 | T-A | HET | INTERGENIC | |
| SBS | Chr6 | 21489093 | T-A | HET | INTERGENIC | |
| SBS | Chr6 | 26858058 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os06g44460 |
| SBS | Chr6 | 615166 | C-G | HOMO | INTERGENIC | |
| SBS | Chr6 | 6959951 | C-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 1147953 | C-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 12891480 | C-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 28600893 | C-T | HOMO | INTERGENIC | |
| SBS | Chr8 | 12637664 | G-C | HOMO | NON_SYNONYMOUS_CODING | LOC_Os08g21140 |
| SBS | Chr8 | 16264354 | G-A | HOMO | INTRON | |
| SBS | Chr8 | 18808807 | C-T | HOMO | INTRON | |
| SBS | Chr8 | 9191549 | C-T | HOMO | INTERGENIC | |
| SBS | Chr9 | 4076991 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr9 | 4199948 | G-C | HET | INTERGENIC | |
| SBS | Chr9 | 5709653 | G-C | HET | INTERGENIC | |
| SBS | Chr9 | 7260839 | G-A | HOMO | INTERGENIC |
Deletions: 30
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr5 | 3072163 | 3072164 | 1 | |
| Deletion | Chr3 | 4052381 | 4052382 | 1 | |
| Deletion | Chr7 | 4255529 | 4255532 | 3 | |
| Deletion | Chr8 | 4433145 | 4433154 | 9 | |
| Deletion | Chr4 | 5577829 | 5577834 | 5 | |
| Deletion | Chr7 | 8388856 | 8388858 | 2 | |
| Deletion | Chr5 | 9109445 | 9109447 | 2 | LOC_Os05g16110 |
| Deletion | Chr5 | 9232879 | 9232886 | 7 | LOC_Os05g16300 |
| Deletion | Chr3 | 9472945 | 9472946 | 1 | |
| Deletion | Chr8 | 9660444 | 9660450 | 6 | |
| Deletion | Chr8 | 9667223 | 9667224 | 1 | |
| Deletion | Chr5 | 9679274 | 9679275 | 1 | |
| Deletion | Chr8 | 10546350 | 10546351 | 1 | |
| Deletion | Chr9 | 15948262 | 15948263 | 1 | |
| Deletion | Chr6 | 19781161 | 19781162 | 1 | LOC_Os06g33970 |
| Deletion | Chr4 | 20350384 | 20350390 | 6 | |
| Deletion | Chr12 | 20782912 | 20782923 | 11 | |
| Deletion | Chr4 | 20933866 | 20933910 | 44 | |
| Deletion | Chr4 | 21159541 | 21159544 | 3 | |
| Deletion | Chr10 | 21754398 | 21769514 | 15116 | 2 |
| Deletion | Chr8 | 22026223 | 22026230 | 7 | |
| Deletion | Chr9 | 22185518 | 22185519 | 1 | |
| Deletion | Chr3 | 22542587 | 22542588 | 1 | |
| Deletion | Chr4 | 23750601 | 23750606 | 5 | LOC_Os04g39880 |
| Deletion | Chr1 | 25353737 | 25353739 | 2 | |
| Deletion | Chr6 | 26024043 | 26024044 | 1 | |
| Deletion | Chr6 | 26857814 | 26857820 | 6 | |
| Deletion | Chr5 | 27596623 | 27596624 | 1 | |
| Deletion | Chr4 | 30608633 | 30608642 | 9 | |
| Deletion | Chr1 | 35868650 | 35868662 | 12 |
Insertions: 39
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr6 | 13325345 | 14791776 | |
| Inversion | Chr7 | 20149467 | 20385976 | LOC_Os07g33710 |
No Translocation
