Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN-76 [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN-76 Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 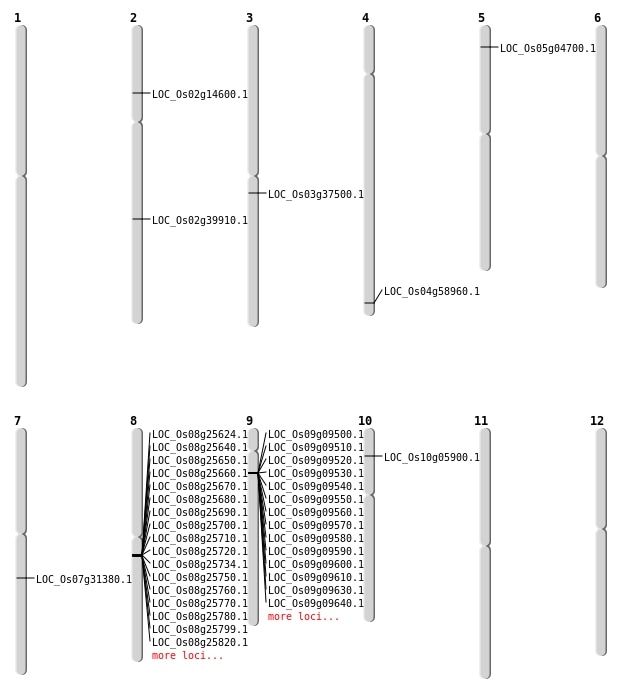
|
| Variant Information |
|---|
Single base substitutions: 31
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 17833819 | G-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 20335370 | C-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 21387470 | C-T | HET | INTRON | |
| SBS | Chr10 | 1102960 | G-A | HOMO | INTERGENIC | |
| SBS | Chr11 | 10971927 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 2441486 | C-T | HOMO | INTERGENIC | |
| SBS | Chr11 | 2458479 | A-C | HOMO | INTERGENIC | |
| SBS | Chr11 | 27238381 | C-T | HET | INTRON | |
| SBS | Chr11 | 8823172 | C-T | HOMO | INTERGENIC | |
| SBS | Chr12 | 10271759 | G-T | HOMO | INTERGENIC | |
| SBS | Chr12 | 22170139 | G-T | HOMO | INTERGENIC | |
| SBS | Chr12 | 4867879 | G-C | HET | INTRON | |
| SBS | Chr2 | 28085258 | T-C | HET | INTERGENIC | |
| SBS | Chr2 | 32104422 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 8059331 | A-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os02g14600 |
| SBS | Chr2 | 8059332 | C-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr3 | 13597665 | A-C | HOMO | INTERGENIC | |
| SBS | Chr3 | 22376541 | G-A | HOMO | INTRON | |
| SBS | Chr4 | 11598263 | G-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 16222604 | A-T | HOMO | INTERGENIC | |
| SBS | Chr4 | 19777016 | G-C | HOMO | INTERGENIC | |
| SBS | Chr4 | 34601586 | A-G | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr4 | 8097595 | C-T | HOMO | INTERGENIC | |
| SBS | Chr5 | 15785436 | A-G | HET | INTERGENIC | |
| SBS | Chr5 | 17678705 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 16196322 | A-T | HET | INTERGENIC | |
| SBS | Chr6 | 6181979 | C-G | HOMO | INTERGENIC | |
| SBS | Chr7 | 18552720 | G-A | HET | INTRON | |
| SBS | Chr7 | 229861 | T-C | HET | INTERGENIC | |
| SBS | Chr8 | 11503609 | A-C | HET | INTERGENIC | |
| SBS | Chr9 | 5495450 | G-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os09g10080 |
Deletions: 19
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr12 | 154850 | 154851 | 1 | |
| Deletion | Chr5 | 2219950 | 2219972 | 22 | LOC_Os05g04700 |
| Deletion | Chr9 | 5120001 | 5354000 | 234000 | 34 |
| Deletion | Chr9 | 5365001 | 5491000 | 126000 | 18 |
| Deletion | Chr4 | 10836324 | 10836334 | 10 | |
| Deletion | Chr11 | 11095612 | 11095613 | 1 | |
| Deletion | Chr7 | 11344846 | 11344852 | 6 | |
| Deletion | Chr2 | 11872851 | 11872862 | 11 | |
| Deletion | Chr1 | 12874521 | 12874523 | 2 | |
| Deletion | Chr8 | 15591001 | 15723000 | 132000 | 18 |
| Deletion | Chr9 | 15840303 | 15840308 | 5 | |
| Deletion | Chr9 | 17369080 | 17369083 | 3 | |
| Deletion | Chr5 | 20019794 | 20019798 | 4 | |
| Deletion | Chr3 | 20808148 | 20808149 | 1 | LOC_Os03g37500 |
| Deletion | Chr11 | 23164943 | 23164944 | 1 | |
| Deletion | Chr2 | 24133854 | 24133856 | 2 | LOC_Os02g39910 |
| Deletion | Chr6 | 25232498 | 25232508 | 10 | |
| Deletion | Chr4 | 28851992 | 28851996 | 4 | |
| Deletion | Chr4 | 35074674 | 35074677 | 3 | LOC_Os04g58960 |
Insertions: 1
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr11 | 25148595 | 25148595 | 1 |
Inversions: 3
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr9 | 5596177 | 5785805 | LOC_Os09g10600 |
| Inversion | Chr9 | 5596190 | 5785802 | LOC_Os09g10600 |
| Inversion | Chr10 | 12952892 | 13531706 |
Translocations: 5
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr10 | 2979982 | Chr8 | 15590118 | LOC_Os10g05900 |
| Translocation | Chr10 | 12952828 | Chr7 | 18591028 | LOC_Os07g31380 |
| Translocation | Chr10 | 13531701 | Chr7 | 18549307 | |
| Translocation | Chr8 | 15590120 | Chr6 | 24834795 | LOC_Os06g41440 |
| Translocation | Chr8 | 15591599 | Chr6 | 24834791 | LOC_Os06g41440 |
