Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1009-S [Download] |
| Generation | M2-SOS |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1009-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 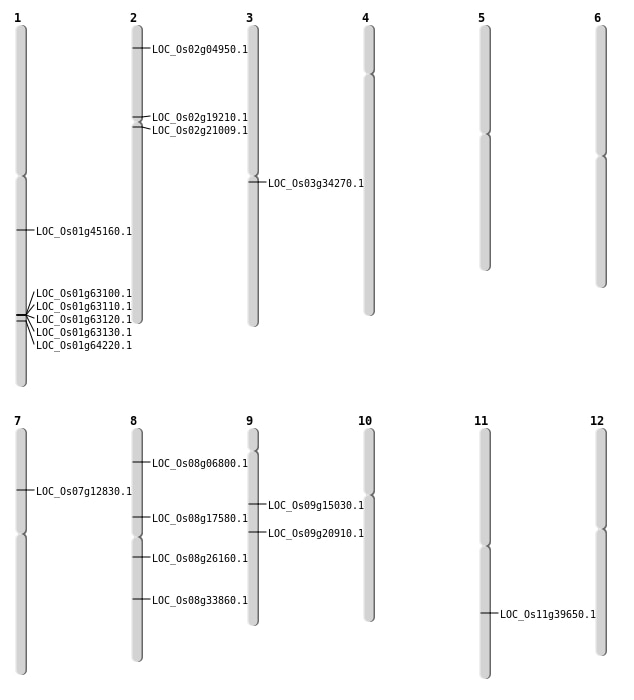
|
| Variant Information |
|---|
Single base substitutions: 32
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 18273899 | C-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr1 | 28414298 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 28414299 | T-G | HET | INTERGENIC | |
| SBS | Chr1 | 28414302 | A-G | HET | INTERGENIC | |
| SBS | Chr1 | 31566848 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 36414607 | G-A | HET | UTR_3_PRIME | |
| SBS | Chr1 | 37295042 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g64220 |
| SBS | Chr1 | 41124404 | A-G | HET | INTERGENIC | |
| SBS | Chr10 | 1823737 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 17095490 | G-T | HET | INTERGENIC | |
| SBS | Chr11 | 21099731 | A-C | HET | INTRON | |
| SBS | Chr11 | 22806061 | T-C | HET | INTERGENIC | |
| SBS | Chr11 | 26178700 | T-C | HET | INTERGENIC | |
| SBS | Chr12 | 2333492 | A-G | HET | INTERGENIC | |
| SBS | Chr2 | 14430870 | A-G | HET | INTERGENIC | |
| SBS | Chr2 | 19051441 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 24099934 | C-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 2855123 | T-C | HOMO | INTERGENIC | |
| SBS | Chr3 | 10093020 | C-A | HET | INTRON | |
| SBS | Chr3 | 19508008 | T-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os03g34270 |
| SBS | Chr3 | 19508009 | T-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os03g34270 |
| SBS | Chr3 | 257974 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 257975 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 6438104 | T-A | HET | INTRON | |
| SBS | Chr6 | 16816900 | A-G | HET | INTERGENIC | |
| SBS | Chr6 | 20158095 | T-G | HET | INTERGENIC | |
| SBS | Chr6 | 27206777 | G-T | HET | INTERGENIC | |
| SBS | Chr6 | 27363484 | T-G | HET | INTERGENIC | |
| SBS | Chr7 | 16458710 | T-A | HET | INTERGENIC | |
| SBS | Chr8 | 15915599 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os08g26160 |
| SBS | Chr9 | 11877286 | T-C | HET | INTERGENIC | |
| SBS | Chr9 | 12022543 | T-C | HET | SYNONYMOUS_CODING |
Deletions: 20
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr2 | 2182298 | 2182435 | 137 | |
| Deletion | Chr2 | 2321164 | 2321165 | 1 | LOC_Os02g04950 |
| Deletion | Chr8 | 3793765 | 3793771 | 6 | LOC_Os08g06800 |
| Deletion | Chr3 | 5996541 | 5996547 | 6 | |
| Deletion | Chr5 | 8928669 | 8928672 | 3 | |
| Deletion | Chr9 | 9068247 | 9068277 | 30 | LOC_Os09g15030 |
| Deletion | Chr8 | 9193389 | 9193404 | 15 | |
| Deletion | Chr2 | 12431001 | 12449000 | 18000 | LOC_Os02g21009 |
| Deletion | Chr9 | 12589423 | 12589440 | 17 | LOC_Os09g20910 |
| Deletion | Chr5 | 16209155 | 16209156 | 1 | |
| Deletion | Chr8 | 21199224 | 21199226 | 2 | LOC_Os08g33860 |
| Deletion | Chr12 | 23467818 | 23467822 | 4 | |
| Deletion | Chr11 | 23615106 | 23615111 | 5 | LOC_Os11g39650 |
| Deletion | Chr7 | 23868064 | 23868073 | 9 | |
| Deletion | Chr1 | 25627280 | 25627295 | 15 | LOC_Os01g45160 |
| Deletion | Chr7 | 27500144 | 27500150 | 6 | |
| Deletion | Chr1 | 28186722 | 28186757 | 35 | |
| Deletion | Chr6 | 29585347 | 29586992 | 1645 | |
| Deletion | Chr1 | 33769682 | 33769707 | 25 | |
| Deletion | Chr1 | 36567001 | 36581000 | 14000 | 4 |
Insertions: 3
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr2 | 29433436 | 29433445 | 10 | |
| Insertion | Chr3 | 24967324 | 24967325 | 2 | |
| Insertion | Chr7 | 11740182 | 11740183 | 2 |
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr2 | 11201962 | 12430983 | LOC_Os02g19210 |
Translocations: 9
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 6993248 | Chr3 | 22046065 | |
| Translocation | Chr7 | 7351923 | Chr2 | 11201969 | LOC_Os02g19210 |
| Translocation | Chr8 | 10754436 | Chr6 | 19260584 | LOC_Os08g17580 |
| Translocation | Chr9 | 15056238 | Chr7 | 1989818 | |
| Translocation | Chr4 | 16244168 | Chr1 | 36566484 | LOC_Os01g63100 |
| Translocation | Chr9 | 22082503 | Chr7 | 7349460 | LOC_Os07g12830 |
| Translocation | Chr9 | 22082545 | Chr6 | 29585382 | |
| Translocation | Chr12 | 25663594 | Chr1 | 29500057 | |
| Translocation | Chr6 | 29586963 | Chr2 | 12449279 | LOC_Os02g21009 |
