Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1011-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1011-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 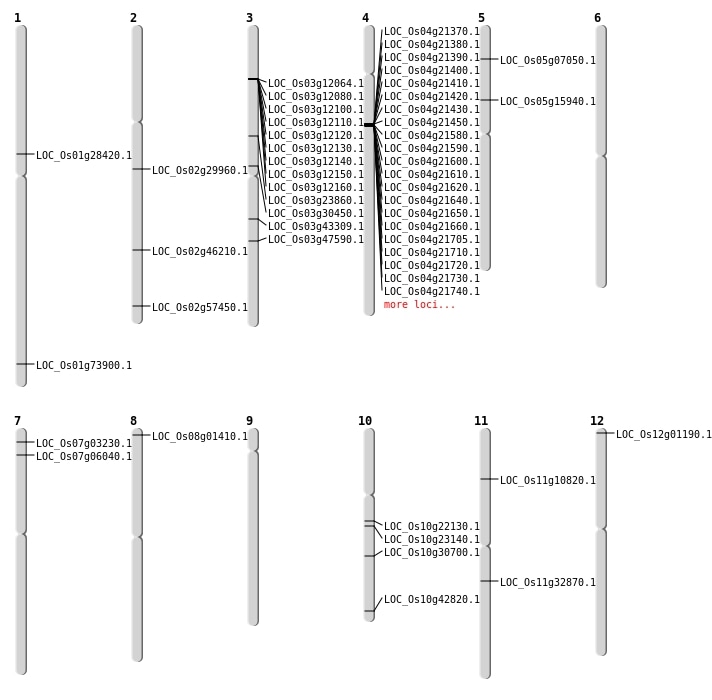
|
| Variant Information |
|---|
Single base substitutions: 33
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 11503172 | C-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 15923518 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os01g28420 |
| SBS | Chr1 | 521630 | T-A | HET | INTRON | |
| SBS | Chr10 | 11429308 | T-G | HET | NON_SYNONYMOUS_CODING | LOC_Os10g22130 |
| SBS | Chr10 | 11709474 | A-C | HET | INTRON | |
| SBS | Chr10 | 16751302 | T-C | HOMO | INTRON | |
| SBS | Chr10 | 23089656 | C-G | HOMO | NON_SYNONYMOUS_CODING | LOC_Os10g42820 |
| SBS | Chr11 | 24237317 | T-A | HET | INTERGENIC | |
| SBS | Chr11 | 26316988 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 5964888 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os11g10820 |
| SBS | Chr12 | 21096316 | T-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr12 | 23269661 | C-T | HET | INTRON | |
| SBS | Chr12 | 23269662 | G-T | HET | INTRON | |
| SBS | Chr12 | 26589654 | C-A | HET | INTERGENIC | |
| SBS | Chr2 | 12011970 | C-T | HET | INTRON | |
| SBS | Chr2 | 21012830 | G-T | HET | INTERGENIC | |
| SBS | Chr2 | 9473480 | A-T | HET | INTERGENIC | |
| SBS | Chr3 | 12333606 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 13542021 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g23860 |
| SBS | Chr3 | 17366868 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os03g30450 |
| SBS | Chr3 | 23677737 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 26937352 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os03g47590 |
| SBS | Chr4 | 20427030 | T-C | HET | UTR_3_PRIME | |
| SBS | Chr5 | 3553076 | T-A | HET | INTERGENIC | |
| SBS | Chr6 | 29214001 | T-A | HET | INTERGENIC | |
| SBS | Chr6 | 30143421 | T-C | HET | INTERGENIC | |
| SBS | Chr7 | 12609176 | A-T | HET | INTRON | |
| SBS | Chr7 | 5217456 | A-C | HET | INTERGENIC | |
| SBS | Chr7 | 7469308 | T-C | HET | INTERGENIC | |
| SBS | Chr8 | 14882864 | C-G | HET | INTERGENIC | |
| SBS | Chr8 | 270186 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os08g01410 |
| SBS | Chr9 | 13586449 | C-T | HET | INTRON | |
| SBS | Chr9 | 13786841 | C-T | HET | INTERGENIC |
Deletions: 27
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr7 | 2931090 | 2931095 | 5 | LOC_Os07g06040 |
| Deletion | Chr5 | 3708714 | 3708716 | 2 | LOC_Os05g07050 |
| Deletion | Chr11 | 4658313 | 4658314 | 1 | |
| Deletion | Chr5 | 6041918 | 6041921 | 3 | |
| Deletion | Chr3 | 6331001 | 6390000 | 59000 | 9 |
| Deletion | Chr7 | 8984896 | 8984897 | 1 | |
| Deletion | Chr5 | 11281510 | 11281511 | 1 | |
| Deletion | Chr4 | 11838762 | 11838768 | 6 | |
| Deletion | Chr4 | 12068001 | 12136000 | 68000 | 8 |
| Deletion | Chr10 | 12081114 | 12081115 | 1 | LOC_Os10g23140 |
| Deletion | Chr2 | 12131359 | 12131365 | 6 | |
| Deletion | Chr4 | 12209001 | 12260000 | 51000 | 8 |
| Deletion | Chr4 | 12276001 | 12870000 | 594000 | 98 |
| Deletion | Chr9 | 12634184 | 12634214 | 30 | |
| Deletion | Chr2 | 13357611 | 13357618 | 7 | |
| Deletion | Chr8 | 16922091 | 16922092 | 1 | |
| Deletion | Chr3 | 17515949 | 17515972 | 23 | |
| Deletion | Chr9 | 19954871 | 19954906 | 35 | |
| Deletion | Chr4 | 21830849 | 21830850 | 1 | |
| Deletion | Chr10 | 21963548 | 21963549 | 1 | |
| Deletion | Chr4 | 22230159 | 22230163 | 4 | LOC_Os04g36820 |
| Deletion | Chr3 | 24144182 | 24145406 | 1224 | LOC_Os03g43309 |
| Deletion | Chr7 | 24586490 | 24586491 | 1 | |
| Deletion | Chr4 | 24868836 | 24868837 | 1 | |
| Deletion | Chr3 | 34034271 | 34034272 | 1 | |
| Deletion | Chr2 | 35209044 | 35209047 | 3 | LOC_Os02g57450 |
| Deletion | Chr1 | 42821538 | 42821592 | 54 | LOC_Os01g73900 |
Insertions: 8
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 12717709 | 12717709 | 1 | |
| Insertion | Chr10 | 22580314 | 22580314 | 1 | |
| Insertion | Chr11 | 10895601 | 10895601 | 1 | |
| Insertion | Chr2 | 15620871 | 15620872 | 2 | |
| Insertion | Chr2 | 17808900 | 17808913 | 14 | LOC_Os02g29960 |
| Insertion | Chr2 | 18371365 | 18371366 | 2 | |
| Insertion | Chr3 | 32604039 | 32604039 | 1 | |
| Insertion | Chr7 | 1274621 | 1274644 | 24 | LOC_Os07g03230 |
Inversions: 5
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr11 | 9134393 | 27265402 | |
| Inversion | Chr4 | 12067553 | 12209189 | LOC_Os04g21580 |
| Inversion | Chr4 | 12135749 | 12870112 | |
| Inversion | Chr10 | 15749530 | 15990474 | LOC_Os10g30700 |
| Inversion | Chr4 | 20084608 | 20761084 | LOC_Os04g33200 |
Translocations: 10
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 105977 | Chr5 | 8998625 | 2 |
| Translocation | Chr11 | 358277 | Chr5 | 8997056 | LOC_Os05g15940 |
| Translocation | Chr5 | 8997053 | Chr4 | 20760808 | LOC_Os05g15940 |
| Translocation | Chr5 | 8997088 | Chr2 | 28153189 | LOC_Os05g15940 |
| Translocation | Chr5 | 8998595 | Chr4 | 20761118 | LOC_Os05g15940 |
| Translocation | Chr4 | 12072508 | Chr2 | 28173309 | |
| Translocation | Chr10 | 15749532 | Chr2 | 28153757 | LOC_Os02g46210 |
| Translocation | Chr10 | 15990461 | Chr3 | 24144186 | 2 |
| Translocation | Chr11 | 19419471 | Chr2 | 28171241 | LOC_Os11g32870 |
| Translocation | Chr4 | 20761074 | Chr2 | 28153141 |
