Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1012-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1012-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 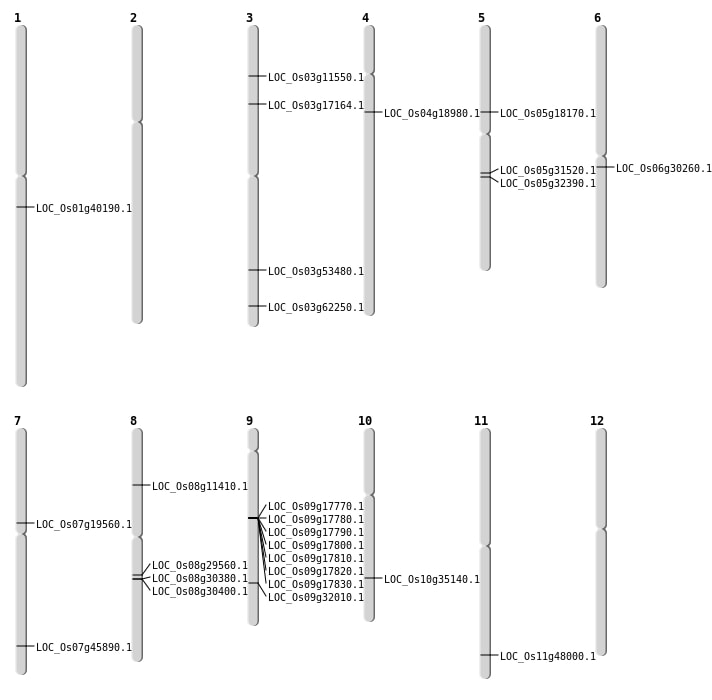
|
| Variant Information |
|---|
Single base substitutions: 36
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 12444464 | A-T | HET | INTERGENIC | |
| SBS | Chr1 | 16095845 | A-G | HOMO | INTERGENIC | |
| SBS | Chr1 | 21018144 | T-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr1 | 38671582 | T-C | HET | INTERGENIC | |
| SBS | Chr1 | 41379019 | C-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 41687767 | G-A | HOMO | INTERGENIC | |
| SBS | Chr10 | 11977648 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 20070624 | A-C | HOMO | INTERGENIC | |
| SBS | Chr10 | 9450421 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr11 | 3773907 | G-T | HOMO | INTERGENIC | |
| SBS | Chr12 | 10237543 | A-G | HET | INTRON | |
| SBS | Chr12 | 13974325 | A-G | HET | INTERGENIC | |
| SBS | Chr12 | 14099264 | C-A | HET | INTERGENIC | |
| SBS | Chr12 | 17125072 | T-A | HET | INTERGENIC | |
| SBS | Chr2 | 14962602 | G-A | HET | UTR_3_PRIME | |
| SBS | Chr3 | 35256165 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os03g62250 |
| SBS | Chr3 | 5338217 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 9547301 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g17164 |
| SBS | Chr4 | 10549502 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g18980 |
| SBS | Chr4 | 25845036 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 9881032 | G-T | HET | INTERGENIC | |
| SBS | Chr5 | 10446800 | T-C | HOMO | NON_SYNONYMOUS_CODING | LOC_Os05g18170 |
| SBS | Chr5 | 29265875 | A-G | HET | INTERGENIC | |
| SBS | Chr5 | 5370435 | G-T | HOMO | INTERGENIC | |
| SBS | Chr5 | 5370436 | C-T | HOMO | INTERGENIC | |
| SBS | Chr6 | 31037671 | A-G | HET | INTRON | |
| SBS | Chr7 | 14816199 | T-A | HET | INTERGENIC | |
| SBS | Chr7 | 24214659 | T-A | HET | INTERGENIC | |
| SBS | Chr7 | 26478686 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 27381689 | T-A | HET | STOP_GAINED | LOC_Os07g45890 |
| SBS | Chr7 | 29526071 | C-G | HET | SYNONYMOUS_CODING | |
| SBS | Chr7 | 4578260 | T-C | HET | INTERGENIC | |
| SBS | Chr7 | 8829323 | G-A | HET | INTRON | |
| SBS | Chr8 | 9671920 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 11324131 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 19550956 | A-C | HET | INTRON |
Deletions: 21
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr1 | 1296206 | 1296209 | 3 | |
| Deletion | Chr11 | 5979033 | 5979077 | 44 | |
| Deletion | Chr3 | 5984070 | 5984073 | 3 | LOC_Os03g11550 |
| Deletion | Chr7 | 8555057 | 8555062 | 5 | |
| Deletion | Chr1 | 8705141 | 8705146 | 5 | |
| Deletion | Chr6 | 9999173 | 9999176 | 3 | |
| Deletion | Chr7 | 10851001 | 10851010 | 9 | |
| Deletion | Chr9 | 10877001 | 10907000 | 30000 | 7 |
| Deletion | Chr7 | 11592778 | 11592779 | 1 | LOC_Os07g19560 |
| Deletion | Chr9 | 11852192 | 11852193 | 1 | |
| Deletion | Chr6 | 17462121 | 17462122 | 1 | LOC_Os06g30260 |
| Deletion | Chr8 | 18690001 | 18705000 | 15000 | 2 |
| Deletion | Chr9 | 19106399 | 19106941 | 542 | LOC_Os09g32010 |
| Deletion | Chr5 | 20124890 | 20124934 | 44 | |
| Deletion | Chr2 | 21367123 | 21367134 | 11 | |
| Deletion | Chr8 | 22280341 | 22280357 | 16 | |
| Deletion | Chr1 | 22672567 | 22677285 | 4718 | LOC_Os01g40190 |
| Deletion | Chr1 | 26029425 | 26029433 | 8 | |
| Deletion | Chr6 | 26076373 | 26076383 | 10 | |
| Deletion | Chr1 | 29026773 | 29026786 | 13 | |
| Deletion | Chr1 | 29539644 | 29539662 | 18 |
Insertions: 3
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr11 | 28676529 | 28676531 | 3 | |
| Insertion | Chr3 | 12114390 | 12114391 | 2 | |
| Insertion | Chr7 | 6446576 | 6446576 | 1 |
Inversions: 5
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr8 | 18122416 | 18690339 | LOC_Os08g29560 |
| Inversion | Chr8 | 18122430 | 18704458 | LOC_Os08g29560 |
| Inversion | Chr5 | 18334747 | 18897509 | 2 |
| Inversion | Chr10 | 18637094 | 18752777 | LOC_Os10g35140 |
| Inversion | Chr11 | 28694608 | 28947547 | LOC_Os11g48000 |
Translocations: 6
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr8 | 6695869 | Chr3 | 30668184 | 2 |
| Translocation | Chr8 | 6695922 | Chr3 | 30668741 | 2 |
| Translocation | Chr8 | 9661127 | Chr5 | 4928416 | |
| Translocation | Chr8 | 11689899 | Chr6 | 17726556 | |
| Translocation | Chr12 | 13380065 | Chr3 | 13641929 | |
| Translocation | Chr9 | 19106941 | Chr1 | 41359779 | LOC_Os09g32010 |
