Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1014-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1014-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 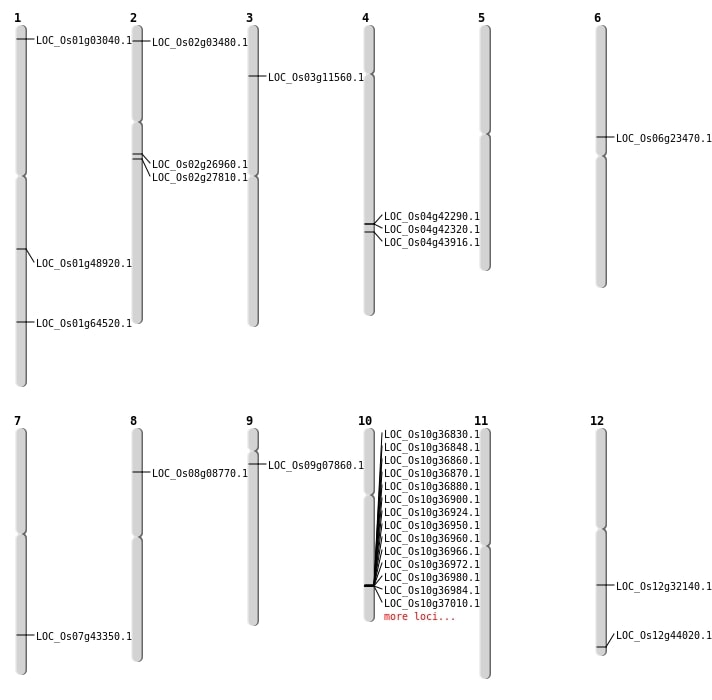
|
| Variant Information |
|---|
Single base substitutions: 39
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 16965235 | G-A | HET | INTRON | |
| SBS | Chr1 | 27206905 | T-A | HOMO | INTERGENIC | |
| SBS | Chr1 | 41522719 | A-G | HET | INTERGENIC | |
| SBS | Chr10 | 14474677 | C-T | HOMO | INTERGENIC | |
| SBS | Chr10 | 18286881 | T-A | HET | INTERGENIC | |
| SBS | Chr10 | 18286882 | C-G | HET | INTERGENIC | |
| SBS | Chr10 | 18286883 | G-C | HET | INTERGENIC | |
| SBS | Chr10 | 20692777 | G-A | HET | UTR_3_PRIME | |
| SBS | Chr11 | 11766399 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 18838613 | G-A | HOMO | INTERGENIC | |
| SBS | Chr11 | 21305594 | C-A | HET | INTERGENIC | |
| SBS | Chr12 | 10595698 | C-T | HOMO | INTERGENIC | |
| SBS | Chr12 | 19385832 | A-G | HOMO | NON_SYNONYMOUS_CODING | LOC_Os12g32140 |
| SBS | Chr12 | 7763454 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr12 | 7984495 | A-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 22270283 | T-A | HET | INTERGENIC | |
| SBS | Chr3 | 30157618 | C-T | HOMO | INTRON | |
| SBS | Chr4 | 27871534 | C-T | HOMO | INTRON | |
| SBS | Chr4 | 9969432 | G-T | HET | INTERGENIC | |
| SBS | Chr5 | 20622354 | C-A | HET | INTRON | |
| SBS | Chr5 | 8291094 | A-G | HET | INTERGENIC | |
| SBS | Chr7 | 12403177 | C-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 17439537 | G-T | HET | INTERGENIC | |
| SBS | Chr7 | 25946572 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os07g43350 |
| SBS | Chr7 | 25974917 | T-C | HET | INTERGENIC | |
| SBS | Chr7 | 26844200 | G-A | HET | INTERGENIC | |
| SBS | Chr8 | 18427974 | G-T | HET | INTERGENIC | |
| SBS | Chr8 | 18427975 | A-G | HET | INTERGENIC | |
| SBS | Chr8 | 24826593 | A-T | HOMO | INTERGENIC | |
| SBS | Chr8 | 24848780 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 27024585 | A-T | HET | INTRON | |
| SBS | Chr8 | 27024586 | A-G | HET | INTRON | |
| SBS | Chr8 | 27596037 | C-T | HET | INTRON | |
| SBS | Chr8 | 5085704 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os08g08770 |
| SBS | Chr8 | 7521483 | A-G | HET | INTERGENIC | |
| SBS | Chr9 | 12972987 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 14118787 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 14588769 | T-G | HOMO | INTERGENIC | |
| SBS | Chr9 | 3997712 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os09g07860 |
Deletions: 23
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr2 | 617518 | 617527 | 9 | |
| Deletion | Chr12 | 2202219 | 2202221 | 2 | |
| Deletion | Chr5 | 2212063 | 2212067 | 4 | |
| Deletion | Chr2 | 3385198 | 3385226 | 28 | |
| Deletion | Chr12 | 4139613 | 4142256 | 2643 | |
| Deletion | Chr8 | 7329519 | 7329528 | 9 | |
| Deletion | Chr7 | 7977387 | 7977404 | 17 | |
| Deletion | Chr1 | 9587570 | 9589347 | 1777 | |
| Deletion | Chr11 | 9804280 | 9804283 | 3 | |
| Deletion | Chr3 | 11044080 | 11044082 | 2 | |
| Deletion | Chr6 | 12531190 | 12531199 | 9 | |
| Deletion | Chr9 | 13903899 | 13903909 | 10 | |
| Deletion | Chr3 | 17591943 | 17591944 | 1 | |
| Deletion | Chr2 | 19066363 | 19066364 | 1 | |
| Deletion | Chr12 | 19365441 | 19365452 | 11 | |
| Deletion | Chr10 | 19716001 | 19814000 | 98000 | 13 |
| Deletion | Chr10 | 19822001 | 20449000 | 627000 | 99 |
| Deletion | Chr10 | 20488001 | 20523000 | 35000 | 7 |
| Deletion | Chr8 | 20703359 | 20703365 | 6 | |
| Deletion | Chr10 | 21690284 | 21690304 | 20 | LOC_Os10g40510 |
| Deletion | Chr4 | 26026176 | 26026193 | 17 | LOC_Os04g43916 |
| Deletion | Chr12 | 27307030 | 27307036 | 6 | LOC_Os12g44020 |
| Deletion | Chr1 | 37440833 | 37440838 | 5 | LOC_Os01g64520 |
Insertions: 7
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 4897757 | 4897758 | 2 | |
| Insertion | Chr2 | 1417698 | 1417699 | 2 | LOC_Os02g03480 |
| Insertion | Chr2 | 16468454 | 16468457 | 4 | LOC_Os02g27810 |
| Insertion | Chr4 | 32200739 | 32200740 | 2 | |
| Insertion | Chr5 | 14520416 | 14520417 | 2 | |
| Insertion | Chr5 | 21507922 | 21507922 | 1 | |
| Insertion | Chr6 | 29030687 | 29030690 | 4 |
Inversions: 3
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr1 | 1163673 | 1503510 | LOC_Os01g03040 |
| Inversion | Chr6 | 13700247 | 14945527 | LOC_Os06g23470 |
| Inversion | Chr6 | 13755758 | 14945532 |
Translocations: 9
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr3 | 5994456 | Chr2 | 15829655 | 2 |
| Translocation | Chr6 | 8355031 | Chr1 | 9587697 | |
| Translocation | Chr6 | 13700262 | Chr1 | 9589369 | LOC_Os06g23470 |
| Translocation | Chr3 | 14301785 | Chr2 | 15830744 | LOC_Os02g26960 |
| Translocation | Chr3 | 14301793 | Chr2 | 15832701 | LOC_Os02g26960 |
| Translocation | Chr2 | 15829387 | Chr1 | 28071434 | 2 |
| Translocation | Chr2 | 15829650 | Chr1 | 28071433 | 2 |
| Translocation | Chr4 | 25020374 | Chr1 | 1503541 | LOC_Os04g42290 |
| Translocation | Chr4 | 25045355 | Chr1 | 1163669 | 2 |
