Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1020-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1020-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 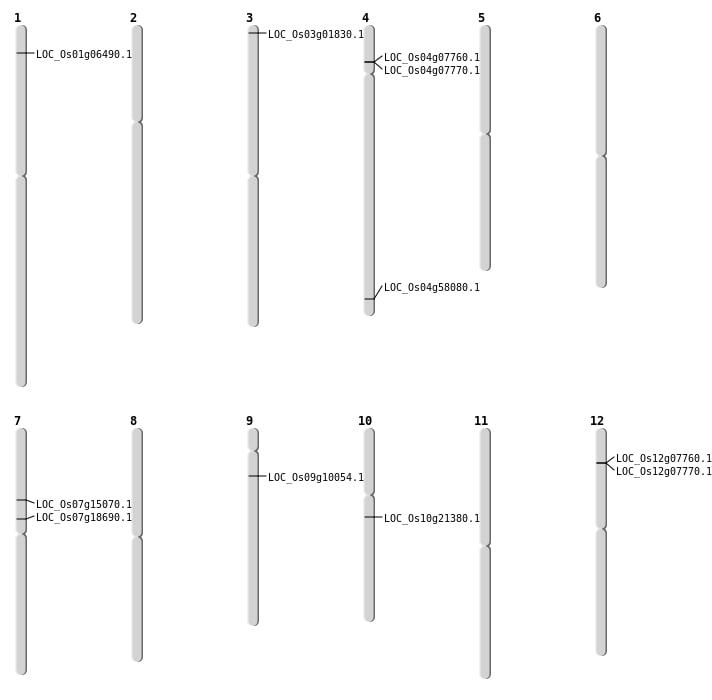
|
| Variant Information |
|---|
Single base substitutions: 39
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 14549903 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 19324357 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 3055733 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g06490 |
| SBS | Chr1 | 38092890 | C-T | HET | INTRON | |
| SBS | Chr1 | 38092891 | C-T | HET | INTRON | |
| SBS | Chr1 | 40041449 | C-G | HET | INTERGENIC | |
| SBS | Chr10 | 9463928 | G-T | HET | INTRON | |
| SBS | Chr10 | 9463930 | G-C | HET | INTRON | |
| SBS | Chr11 | 20632093 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 3967892 | T-C | HOMO | INTERGENIC | |
| SBS | Chr11 | 8270852 | T-G | HOMO | INTERGENIC | |
| SBS | Chr12 | 12470137 | T-C | HET | INTERGENIC | |
| SBS | Chr12 | 2036450 | A-T | HET | INTERGENIC | |
| SBS | Chr2 | 1792468 | G-A | HET | UTR_3_PRIME | |
| SBS | Chr2 | 32359820 | G-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr3 | 22588701 | C-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 23619771 | A-T | HET | INTRON | |
| SBS | Chr3 | 253347 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 25849986 | C-G | HET | INTERGENIC | |
| SBS | Chr4 | 21537121 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 24958089 | G-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 26793711 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 33181647 | A-G | HET | INTERGENIC | |
| SBS | Chr5 | 26369328 | A-T | HET | INTERGENIC | |
| SBS | Chr5 | 26992606 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr5 | 29364868 | T-A | HET | INTERGENIC | |
| SBS | Chr5 | 5490529 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 9196707 | A-G | HET | INTERGENIC | |
| SBS | Chr6 | 22874490 | C-T | HOMO | INTERGENIC | |
| SBS | Chr6 | 8091323 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 11408494 | T-C | HET | INTRON | |
| SBS | Chr7 | 16698171 | C-A | HET | INTERGENIC | |
| SBS | Chr7 | 18940986 | C-A | HET | INTRON | |
| SBS | Chr7 | 26681219 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 8633632 | G-A | HET | STOP_GAINED | LOC_Os07g15070 |
| SBS | Chr8 | 5946526 | T-C | HET | INTRON | |
| SBS | Chr9 | 11654060 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 5822921 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr9 | 6689022 | C-T | HET | INTRON |
Deletions: 22
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr3 | 507396 | 507416 | 20 | LOC_Os03g01830 |
| Deletion | Chr9 | 1287656 | 1287657 | 1 | |
| Deletion | Chr12 | 3903001 | 3919000 | 16000 | 2 |
| Deletion | Chr6 | 4696035 | 4696036 | 1 | |
| Deletion | Chr9 | 5472525 | 5472530 | 5 | LOC_Os09g10054 |
| Deletion | Chr8 | 6262110 | 6262111 | 1 | |
| Deletion | Chr9 | 7170474 | 7170477 | 3 | |
| Deletion | Chr7 | 9846137 | 9846147 | 10 | |
| Deletion | Chr6 | 10095343 | 10095349 | 6 | |
| Deletion | Chr4 | 10581996 | 10581999 | 3 | |
| Deletion | Chr5 | 13281083 | 13281087 | 4 | |
| Deletion | Chr11 | 14693778 | 14693806 | 28 | |
| Deletion | Chr2 | 15968347 | 15968363 | 16 | |
| Deletion | Chr8 | 16629245 | 16629253 | 8 | |
| Deletion | Chr5 | 18192623 | 18192647 | 24 | |
| Deletion | Chr8 | 18716662 | 18716674 | 12 | |
| Deletion | Chr10 | 19927905 | 19927932 | 27 | |
| Deletion | Chr2 | 21527141 | 21527153 | 12 | |
| Deletion | Chr11 | 23139399 | 23139423 | 24 | |
| Deletion | Chr5 | 29364579 | 29364585 | 6 | LOC_Os05g51190 |
| Deletion | Chr3 | 32979463 | 32979468 | 5 | |
| Deletion | Chr4 | 34596457 | 34596552 | 95 | LOC_Os04g58080 |
Insertions: 12
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr11 | 22994407 | 22994409 | 3 | |
| Insertion | Chr11 | 4878367 | 4878368 | 2 | |
| Insertion | Chr12 | 19503149 | 19503150 | 2 | |
| Insertion | Chr12 | 27134457 | 27134460 | 4 | |
| Insertion | Chr2 | 18453732 | 18453739 | 8 | |
| Insertion | Chr2 | 18552323 | 18552324 | 2 | |
| Insertion | Chr4 | 17356573 | 17356574 | 2 | |
| Insertion | Chr4 | 4124944 | 4125002 | 59 | LOC_Os04g07760 |
| Insertion | Chr6 | 10369851 | 10369852 | 2 | |
| Insertion | Chr7 | 11042053 | 11042055 | 3 | LOC_Os07g18690 |
| Insertion | Chr7 | 11351622 | 11351624 | 3 | |
| Insertion | Chr7 | 1455532 | 1455543 | 12 |
Inversions: 3
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr12 | 3893068 | 3919054 | LOC_Os12g07770 |
| Inversion | Chr10 | 15983119 | 15983433 | |
| Inversion | Chr1 | 33332004 | 33332242 |
Translocations: 3
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr4 | 4156867 | Chr2 | 17035193 | LOC_Os04g07770 |
| Translocation | Chr12 | 8885804 | Chr10 | 10726305 | |
| Translocation | Chr10 | 10944016 | Chr2 | 17035207 | LOC_Os10g21380 |
