Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1027-S [Download] |
| Generation | M2-SOS |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1027-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 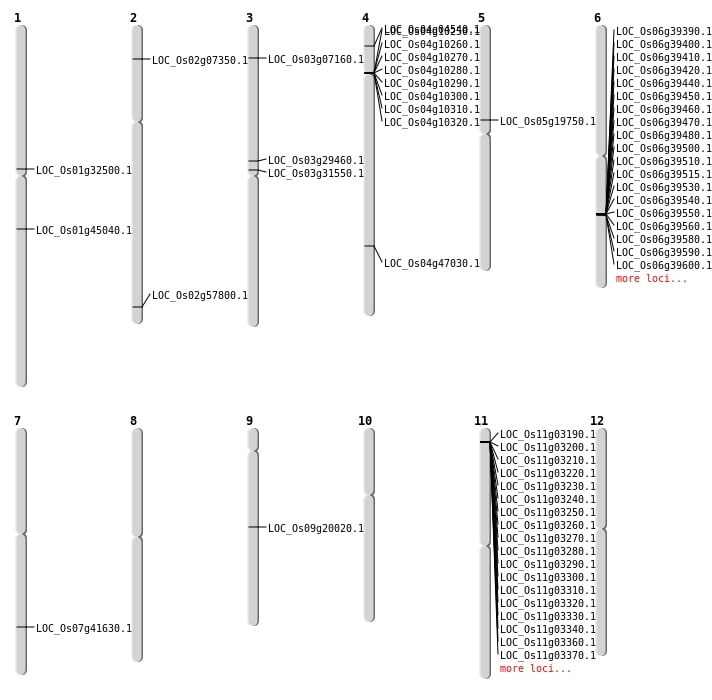
|
| Variant Information |
|---|
Single base substitutions: 35
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 29056360 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 6253233 | C-A | HOMO | UTR_3_PRIME | |
| SBS | Chr1 | 6253234 | T-A | HOMO | UTR_3_PRIME | |
| SBS | Chr10 | 13397231 | C-A | HOMO | INTRON | |
| SBS | Chr11 | 18792327 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 5880468 | G-T | HOMO | INTRON | |
| SBS | Chr11 | 5880469 | C-T | HOMO | INTRON | |
| SBS | Chr2 | 35149462 | A-G | HET | INTERGENIC | |
| SBS | Chr2 | 3602300 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 8636717 | G-C | HET | INTERGENIC | |
| SBS | Chr3 | 20562534 | C-A | HET | INTERGENIC | |
| SBS | Chr3 | 23528387 | G-C | HET | INTERGENIC | |
| SBS | Chr3 | 34939912 | T-C | HOMO | INTRON | |
| SBS | Chr3 | 8902766 | C-T | HOMO | UTR_3_PRIME | |
| SBS | Chr4 | 13247710 | T-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 18753468 | G-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 27102795 | T-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 6330044 | G-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 7690905 | T-A | HET | INTERGENIC | |
| SBS | Chr5 | 11523028 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os05g19750 |
| SBS | Chr5 | 11523029 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os05g19750 |
| SBS | Chr5 | 17227409 | A-T | HET | INTERGENIC | |
| SBS | Chr5 | 17876066 | A-G | HOMO | INTERGENIC | |
| SBS | Chr5 | 25488172 | A-T | HOMO | INTERGENIC | |
| SBS | Chr5 | 27187636 | C-T | HET | INTRON | |
| SBS | Chr5 | 6342472 | T-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr6 | 11189841 | C-G | HET | INTERGENIC | |
| SBS | Chr7 | 16052374 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 24954636 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os07g41630 |
| SBS | Chr7 | 28320185 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 8727107 | C-A | HET | INTERGENIC | |
| SBS | Chr8 | 17671464 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 13838596 | G-A | HET | INTRON | |
| SBS | Chr9 | 15950448 | C-G | HET | INTERGENIC | |
| SBS | Chr9 | 17071614 | T-C | HOMO | INTERGENIC |
Deletions: 30
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr3 | 553769 | 553775 | 6 | |
| Deletion | Chr11 | 1159001 | 1357000 | 198000 | 30 |
| Deletion | Chr2 | 3796758 | 3796770 | 12 | LOC_Os02g07350 |
| Deletion | Chr3 | 4592313 | 4592322 | 9 | |
| Deletion | Chr1 | 4717554 | 4717570 | 16 | |
| Deletion | Chr4 | 5538001 | 5574000 | 36000 | 8 |
| Deletion | Chr2 | 7490201 | 7490202 | 1 | |
| Deletion | Chr12 | 8008909 | 8008924 | 15 | |
| Deletion | Chr12 | 8814885 | 8814899 | 14 | |
| Deletion | Chr1 | 10465315 | 10465316 | 1 | |
| Deletion | Chr9 | 11992159 | 11992162 | 3 | LOC_Os09g20020 |
| Deletion | Chr10 | 13424086 | 13424089 | 3 | |
| Deletion | Chr3 | 16691196 | 16691216 | 20 | |
| Deletion | Chr3 | 16787359 | 16787361 | 2 | LOC_Os03g29460 |
| Deletion | Chr2 | 16798688 | 16798698 | 10 | |
| Deletion | Chr1 | 17823668 | 17823678 | 10 | LOC_Os01g32500 |
| Deletion | Chr2 | 18023904 | 18024048 | 144 | |
| Deletion | Chr3 | 19599574 | 19599586 | 12 | |
| Deletion | Chr2 | 20362083 | 20362091 | 8 | |
| Deletion | Chr8 | 20766241 | 20766245 | 4 | |
| Deletion | Chr8 | 22205703 | 22205704 | 1 | |
| Deletion | Chr7 | 22335438 | 22335470 | 32 | |
| Deletion | Chr2 | 23238307 | 23238327 | 20 | |
| Deletion | Chr6 | 23376001 | 23564000 | 188000 | 28 |
| Deletion | Chr5 | 23866672 | 23866687 | 15 | |
| Deletion | Chr5 | 25488179 | 25488180 | 1 | |
| Deletion | Chr4 | 27873128 | 27873133 | 5 | LOC_Os04g47030 |
| Deletion | Chr11 | 28662360 | 28662365 | 5 | |
| Deletion | Chr2 | 35402901 | 35402907 | 6 | LOC_Os02g57800 |
| Deletion | Chr2 | 35408957 | 35408979 | 22 |
Insertions: 8
No Inversion
Translocations: 6
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 1158337 | Chr3 | 3665909 | LOC_Os03g07160 |
| Translocation | Chr11 | 1158976 | Chr3 | 3665901 | 2 |
| Translocation | Chr4 | 2172575 | Chr1 | 27404759 | LOC_Os04g04540 |
| Translocation | Chr3 | 17986429 | Chr2 | 18023915 | LOC_Os03g31550 |
| Translocation | Chr11 | 21302769 | Chr1 | 25564588 | LOC_Os01g45040 |
| Translocation | Chr8 | 23485349 | Chr6 | 23717186 |
