Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1029-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1029-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 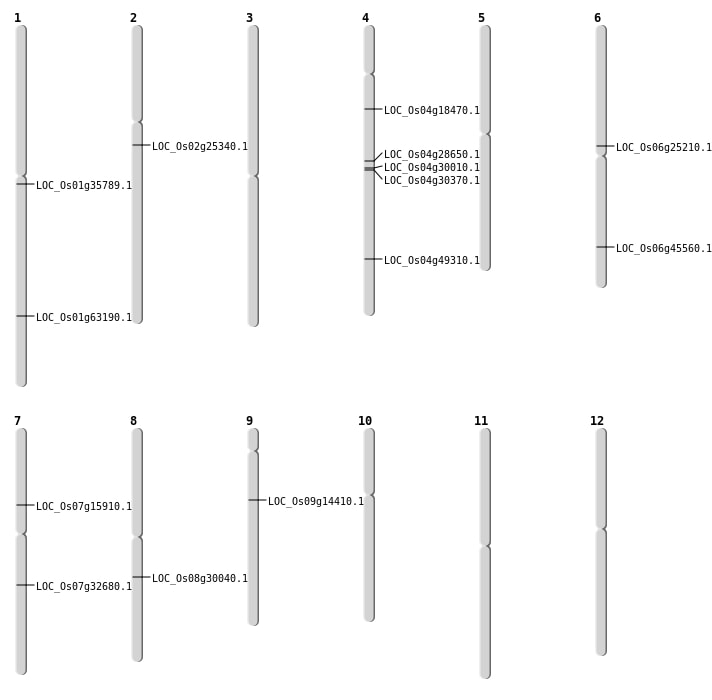
|
| Variant Information |
|---|
Single base substitutions: 40
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 13421198 | C-G | HET | INTERGENIC | |
| SBS | Chr1 | 2005410 | T-A | HET | INTERGENIC | |
| SBS | Chr1 | 25538550 | C-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr1 | 9904929 | G-T | HET | INTERGENIC | |
| SBS | Chr1 | 9925852 | C-A | HET | INTERGENIC | |
| SBS | Chr10 | 1440878 | G-T | HET | INTERGENIC | |
| SBS | Chr11 | 2443941 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 25035356 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 27375804 | G-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr12 | 2785822 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 31644893 | A-G | HET | INTERGENIC | |
| SBS | Chr2 | 8866651 | T-C | HET | INTERGENIC | |
| SBS | Chr3 | 1493516 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 18899016 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 6097040 | A-T | HET | INTERGENIC | |
| SBS | Chr3 | 9158288 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 15036380 | T-C | HET | INTERGENIC | |
| SBS | Chr4 | 16971269 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g28650 |
| SBS | Chr4 | 1844226 | C-G | HOMO | INTRON | |
| SBS | Chr4 | 20418554 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 28273482 | T-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 28273483 | A-G | HOMO | INTERGENIC | |
| SBS | Chr4 | 29434225 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g49310 |
| SBS | Chr4 | 29823954 | A-T | HET | INTERGENIC | |
| SBS | Chr4 | 33184238 | A-G | HOMO | INTERGENIC | |
| SBS | Chr4 | 7425409 | A-G | HET | INTERGENIC | |
| SBS | Chr5 | 29535944 | T-A | HOMO | INTERGENIC | |
| SBS | Chr5 | 5155508 | A-G | HET | INTERGENIC | |
| SBS | Chr5 | 8036050 | G-T | HET | INTRON | |
| SBS | Chr5 | 9352790 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 13963828 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 14712573 | C-A | HET | INTERGENIC | |
| SBS | Chr7 | 19515986 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os07g32680 |
| SBS | Chr7 | 7054866 | T-A | HOMO | INTERGENIC | |
| SBS | Chr7 | 9247049 | G-C | HOMO | NON_SYNONYMOUS_CODING | LOC_Os07g15910 |
| SBS | Chr8 | 16290792 | A-G | HOMO | INTERGENIC | |
| SBS | Chr8 | 18463592 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os08g30040 |
| SBS | Chr8 | 6183119 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 12890251 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 1635409 | C-A | HET | INTERGENIC |
Deletions: 22
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr4 | 4818771 | 4818791 | 20 | |
| Deletion | Chr7 | 7055046 | 7055047 | 1 | |
| Deletion | Chr10 | 7961374 | 7961382 | 8 | |
| Deletion | Chr9 | 8516431 | 8517135 | 704 | LOC_Os09g14410 |
| Deletion | Chr1 | 10473617 | 10473618 | 1 | |
| Deletion | Chr9 | 12417191 | 12417198 | 7 | |
| Deletion | Chr11 | 12616712 | 12616722 | 10 | |
| Deletion | Chr9 | 13592760 | 13592776 | 16 | |
| Deletion | Chr6 | 14753726 | 14753728 | 2 | LOC_Os06g25210 |
| Deletion | Chr2 | 14774500 | 14774501 | 1 | LOC_Os02g25340 |
| Deletion | Chr11 | 15644260 | 15644261 | 1 | |
| Deletion | Chr8 | 16112082 | 16112083 | 1 | |
| Deletion | Chr9 | 18705878 | 18705894 | 16 | |
| Deletion | Chr2 | 19697590 | 19697591 | 1 | |
| Deletion | Chr9 | 22840257 | 22840269 | 12 | |
| Deletion | Chr5 | 25851803 | 25851805 | 2 | |
| Deletion | Chr8 | 26448128 | 26448131 | 3 | |
| Deletion | Chr4 | 27017014 | 27017024 | 10 | |
| Deletion | Chr6 | 27585811 | 27585814 | 3 | LOC_Os06g45560 |
| Deletion | Chr4 | 28031180 | 28031181 | 1 | |
| Deletion | Chr4 | 31906335 | 31906345 | 10 | |
| Deletion | Chr1 | 36626198 | 36626207 | 9 | LOC_Os01g63190 |
Insertions: 3
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 19824972 | 19824989 | 18 | LOC_Os01g35789 |
| Insertion | Chr11 | 22527025 | 22527027 | 3 | |
| Insertion | Chr9 | 10403603 | 10403604 | 2 |
Inversions: 3
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr12 | 12719363 | 13831880 | |
| Inversion | Chr4 | 17906985 | 18125356 | 2 |
| Inversion | Chr5 | 20904919 | 24235903 |
Translocations: 4
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 4838478 | Chr4 | 18124179 | LOC_Os04g30370 |
| Translocation | Chr10 | 10311978 | Chr4 | 5525930 | |
| Translocation | Chr6 | 12594687 | Chr1 | 32531298 | |
| Translocation | Chr11 | 13259843 | Chr4 | 10208284 | LOC_Os04g18470 |
