Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1033-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1033-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 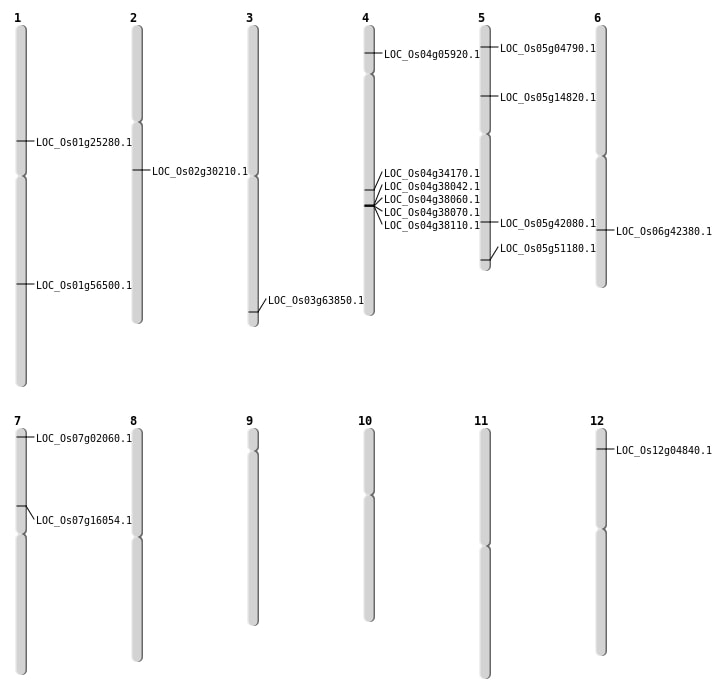
|
| Variant Information |
|---|
Single base substitutions: 35
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 13722673 | A-C | HET | INTERGENIC | |
| SBS | Chr1 | 14281075 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os01g25280 |
| SBS | Chr1 | 16143370 | C-A | HET | INTERGENIC | |
| SBS | Chr1 | 16603406 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr1 | 24513201 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 40211109 | G-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 40218609 | T-C | HOMO | INTERGENIC | |
| SBS | Chr10 | 13812706 | T-G | HET | INTRON | |
| SBS | Chr10 | 13820193 | C-A | HET | INTERGENIC | |
| SBS | Chr10 | 22905633 | G-T | HET | INTERGENIC | |
| SBS | Chr10 | 3134328 | A-T | HET | INTERGENIC | |
| SBS | Chr10 | 4098450 | A-C | HOMO | INTRON | |
| SBS | Chr11 | 19299703 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 2082211 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os12g04840 |
| SBS | Chr12 | 2082218 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os12g04840 |
| SBS | Chr12 | 4705663 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 12887160 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 19332014 | G-T | HET | UTR_5_PRIME | |
| SBS | Chr2 | 19517941 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 35013511 | C-A | HET | INTERGENIC | |
| SBS | Chr2 | 5884939 | A-C | HET | INTERGENIC | |
| SBS | Chr3 | 14901752 | C-T | HET | INTRON | |
| SBS | Chr3 | 26985054 | C-T | HOMO | UTR_3_PRIME | |
| SBS | Chr3 | 27401335 | T-G | HET | INTERGENIC | |
| SBS | Chr4 | 11010425 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 19694684 | T-A | HET | INTERGENIC | |
| SBS | Chr4 | 4888093 | A-G | HOMO | INTERGENIC | |
| SBS | Chr5 | 1825868 | C-A | HET | INTERGENIC | |
| SBS | Chr5 | 2278000 | C-T | HET | STOP_GAINED | LOC_Os05g04790 |
| SBS | Chr5 | 24618642 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os05g42080 |
| SBS | Chr5 | 29358341 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os05g51180 |
| SBS | Chr6 | 24597852 | C-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 26760017 | G-A | HET | INTERGENIC | |
| SBS | Chr8 | 3354782 | A-G | HOMO | INTERGENIC | |
| SBS | Chr8 | 7133483 | C-T | HET | INTERGENIC |
Deletions: 32
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr9 | 192739 | 192754 | 15 | |
| Deletion | Chr7 | 633919 | 633920 | 1 | LOC_Os07g02060 |
| Deletion | Chr9 | 1908231 | 1916646 | 8415 | |
| Deletion | Chr4 | 3064717 | 3064721 | 4 | LOC_Os04g05920 |
| Deletion | Chr10 | 3478370 | 3478371 | 1 | |
| Deletion | Chr4 | 4763548 | 4763551 | 3 | |
| Deletion | Chr9 | 6187456 | 6187458 | 2 | |
| Deletion | Chr1 | 6395986 | 6395987 | 1 | |
| Deletion | Chr3 | 6439786 | 6439787 | 1 | |
| Deletion | Chr4 | 7154216 | 7154223 | 7 | |
| Deletion | Chr1 | 7487572 | 7487576 | 4 | |
| Deletion | Chr7 | 9359689 | 9359690 | 1 | LOC_Os07g16054 |
| Deletion | Chr9 | 9981779 | 9981784 | 5 | |
| Deletion | Chr12 | 10872444 | 10872452 | 8 | |
| Deletion | Chr3 | 11248369 | 11248385 | 16 | |
| Deletion | Chr8 | 12869966 | 12869970 | 4 | |
| Deletion | Chr11 | 13111035 | 13111050 | 15 | |
| Deletion | Chr3 | 13195069 | 13195082 | 13 | |
| Deletion | Chr3 | 14081347 | 14081353 | 6 | |
| Deletion | Chr2 | 17959783 | 17959787 | 4 | LOC_Os02g30210 |
| Deletion | Chr11 | 18013384 | 18013386 | 2 | |
| Deletion | Chr12 | 19500573 | 19500574 | 1 | |
| Deletion | Chr4 | 19877684 | 19877691 | 7 | |
| Deletion | Chr4 | 20709177 | 20709180 | 3 | LOC_Os04g34170 |
| Deletion | Chr8 | 21263899 | 21263901 | 2 | |
| Deletion | Chr4 | 22647001 | 22661000 | 14000 | 3 |
| Deletion | Chr6 | 25474249 | 25474253 | 4 | LOC_Os06g42380 |
| Deletion | Chr3 | 26658306 | 26658314 | 8 | |
| Deletion | Chr5 | 28341507 | 28341509 | 2 | |
| Deletion | Chr1 | 32575100 | 32575105 | 5 | LOC_Os01g56500 |
| Deletion | Chr1 | 34127747 | 34127750 | 3 | |
| Deletion | Chr3 | 36073616 | 36073617 | 1 | LOC_Os03g63850 |
Insertions: 4
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 40218598 | 40218620 | 23 | |
| Insertion | Chr2 | 12864063 | 12864066 | 4 | |
| Insertion | Chr3 | 24678623 | 24678623 | 1 | |
| Insertion | Chr3 | 8453398 | 8453405 | 8 |
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr4 | 22646520 | 22680439 | 2 |
Translocations: 1
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr5 | 8428042 | Chr1 | 23528701 | LOC_Os05g14820 |
