Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1037-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1037-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 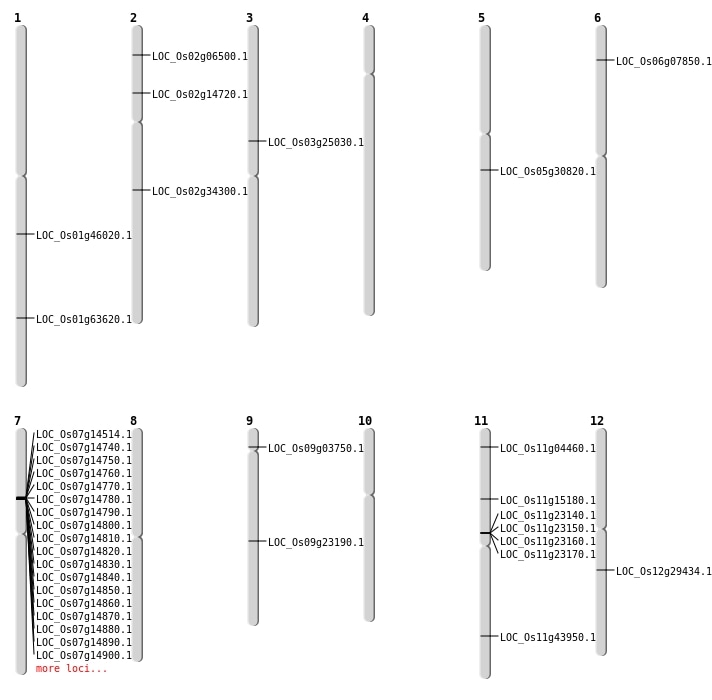
|
| Variant Information |
|---|
Single base substitutions: 35
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 2004765 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 36895447 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr1 | 36895520 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g63620 |
| SBS | Chr10 | 21314922 | T-A | HOMO | INTRON | |
| SBS | Chr10 | 21423634 | T-A | HOMO | INTERGENIC | |
| SBS | Chr11 | 12307368 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 21184481 | C-A | HET | INTERGENIC | |
| SBS | Chr11 | 23204832 | C-T | HOMO | INTERGENIC | |
| SBS | Chr11 | 26545797 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os11g43950 |
| SBS | Chr11 | 8562967 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os11g15180 |
| SBS | Chr11 | 9147620 | A-T | HET | INTRON | |
| SBS | Chr12 | 17486294 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os12g29434 |
| SBS | Chr12 | 25867210 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr2 | 20214860 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 20526845 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os02g34300 |
| SBS | Chr2 | 20761306 | A-T | HET | INTERGENIC | |
| SBS | Chr2 | 3882185 | G-A | HET | INTRON | |
| SBS | Chr3 | 21445420 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 28161928 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 32300670 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 10430228 | G-T | HOMO | INTERGENIC | |
| SBS | Chr4 | 12926076 | G-C | HET | INTERGENIC | |
| SBS | Chr4 | 4805866 | G-T | HOMO | INTERGENIC | |
| SBS | Chr5 | 10616827 | T-C | HET | INTERGENIC | |
| SBS | Chr5 | 17796154 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 18045057 | G-T | HET | INTERGENIC | |
| SBS | Chr5 | 8462694 | C-T | HOMO | INTERGENIC | |
| SBS | Chr6 | 1152088 | T-A | HOMO | INTERGENIC | |
| SBS | Chr6 | 12365908 | T-G | HET | INTERGENIC | |
| SBS | Chr6 | 16661643 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 3280889 | G-A | HOMO | INTERGENIC | |
| SBS | Chr7 | 10488459 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 8030499 | T-C | HET | INTERGENIC | |
| SBS | Chr8 | 17922241 | C-A | HOMO | INTRON | |
| SBS | Chr8 | 4024814 | G-A | HET | INTRON |
Deletions: 26
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr3 | 527413 | 527415 | 2 | |
| Deletion | Chr11 | 1865026 | 1865027 | 1 | LOC_Os11g04460 |
| Deletion | Chr9 | 1897708 | 1899123 | 1415 | LOC_Os09g03750 |
| Deletion | Chr6 | 2214221 | 2214223 | 2 | |
| Deletion | Chr2 | 3238634 | 3238652 | 18 | LOC_Os02g06500 |
| Deletion | Chr2 | 3475858 | 3475870 | 12 | |
| Deletion | Chr9 | 5589738 | 5589741 | 3 | |
| Deletion | Chr8 | 6279228 | 6279230 | 2 | |
| Deletion | Chr7 | 8264043 | 8264076 | 33 | |
| Deletion | Chr7 | 8403001 | 8581000 | 178000 | 22 |
| Deletion | Chr2 | 8870408 | 8870409 | 1 | |
| Deletion | Chr7 | 11160324 | 11160328 | 4 | |
| Deletion | Chr6 | 12666541 | 12666560 | 19 | |
| Deletion | Chr11 | 13313472 | 13333779 | 20307 | 4 |
| Deletion | Chr3 | 14292439 | 14292465 | 26 | LOC_Os03g25030 |
| Deletion | Chr12 | 17486727 | 17486733 | 6 | |
| Deletion | Chr9 | 19048183 | 19048191 | 8 | |
| Deletion | Chr5 | 19464260 | 19464265 | 5 | |
| Deletion | Chr12 | 20728406 | 20728407 | 1 | |
| Deletion | Chr3 | 24027380 | 24027382 | 2 | |
| Deletion | Chr3 | 24342396 | 24342402 | 6 | |
| Deletion | Chr1 | 26163932 | 26163933 | 1 | LOC_Os01g46020 |
| Deletion | Chr6 | 26216914 | 26216934 | 20 | LOC_Os06g43590 |
| Deletion | Chr5 | 26233863 | 26233866 | 3 | |
| Deletion | Chr3 | 31633423 | 31633455 | 32 | |
| Deletion | Chr1 | 35738842 | 35738857 | 15 |
Insertions: 5
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr7 | 8275823 | 8403299 | LOC_Os07g14514 |
Translocations: 6
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr9 | 1897705 | Chr2 | 21866910 | LOC_Os09g03750 |
| Translocation | Chr9 | 1899085 | Chr2 | 21866876 | LOC_Os09g03750 |
| Translocation | Chr6 | 3815256 | Chr5 | 17887274 | 2 |
| Translocation | Chr6 | 3815847 | Chr5 | 17887288 | 2 |
| Translocation | Chr9 | 13753267 | Chr8 | 23868697 | LOC_Os09g23190 |
| Translocation | Chr4 | 17080140 | Chr2 | 8120312 | LOC_Os02g14720 |
