Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1046-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1046-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 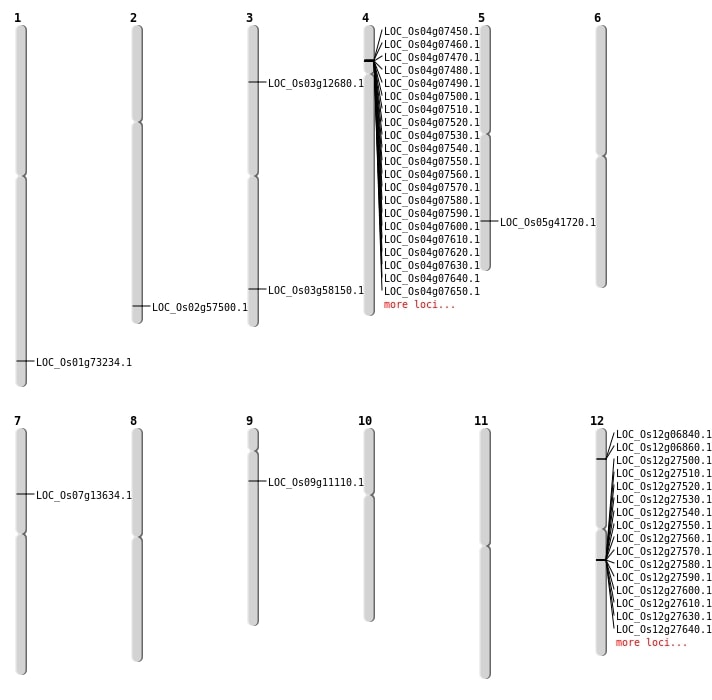
|
| Variant Information |
|---|
Single base substitutions: 27
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 28334529 | A-T | HET | INTERGENIC | |
| SBS | Chr1 | 35796100 | T-A | HET | INTERGENIC | |
| SBS | Chr1 | 40416286 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 9395740 | C-A | HET | INTRON | |
| SBS | Chr10 | 22905633 | G-T | HOMO | INTERGENIC | |
| SBS | Chr12 | 1173294 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr12 | 24429893 | T-C | HOMO | NON_SYNONYMOUS_CODING | LOC_Os12g39590 |
| SBS | Chr12 | 3323532 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g06840 |
| SBS | Chr12 | 3323534 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g06840 |
| SBS | Chr12 | 3335484 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os12g06860 |
| SBS | Chr12 | 4966880 | T-A | HET | INTERGENIC | |
| SBS | Chr2 | 19307946 | A-G | HET | INTERGENIC | |
| SBS | Chr3 | 27940619 | G-C | HET | INTERGENIC | |
| SBS | Chr3 | 6762038 | A-G | HOMO | INTERGENIC | |
| SBS | Chr4 | 10579442 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g19060 |
| SBS | Chr4 | 10634328 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr4 | 1829220 | T-A | HET | INTERGENIC | |
| SBS | Chr4 | 20051518 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os04g33120 |
| SBS | Chr4 | 24901550 | C-G | HET | INTERGENIC | |
| SBS | Chr4 | 8412105 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 1266185 | T-C | HET | INTERGENIC | |
| SBS | Chr7 | 13475601 | A-G | HET | INTERGENIC | |
| SBS | Chr7 | 5951050 | C-A | HET | INTERGENIC | |
| SBS | Chr8 | 23241237 | A-G | HOMO | INTERGENIC | |
| SBS | Chr8 | 23848019 | G-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr9 | 1767768 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 9890749 | G-A | HET | INTERGENIC |
Deletions: 37
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr9 | 728404 | 728411 | 7 | |
| Deletion | Chr5 | 1163124 | 1163145 | 21 | |
| Deletion | Chr7 | 2586780 | 2586785 | 5 | |
| Deletion | Chr3 | 3885185 | 3885186 | 1 | |
| Deletion | Chr4 | 3955001 | 4123000 | 168000 | 31 |
| Deletion | Chr9 | 6161901 | 6161909 | 8 | LOC_Os09g11110 |
| Deletion | Chr1 | 6316525 | 6316526 | 1 | |
| Deletion | Chr3 | 6762838 | 6764306 | 1468 | LOC_Os03g12680 |
| Deletion | Chr7 | 7794563 | 7794568 | 5 | |
| Deletion | Chr7 | 7815604 | 7815605 | 1 | LOC_Os07g13634 |
| Deletion | Chr11 | 8504811 | 8504843 | 32 | |
| Deletion | Chr9 | 9351193 | 9351200 | 7 | |
| Deletion | Chr10 | 9552389 | 9552391 | 2 | |
| Deletion | Chr9 | 9622849 | 9622916 | 67 | |
| Deletion | Chr3 | 11237083 | 11237089 | 6 | |
| Deletion | Chr4 | 11363426 | 11363461 | 35 | |
| Deletion | Chr1 | 13679880 | 13679893 | 13 | |
| Deletion | Chr12 | 14712629 | 14712633 | 4 | |
| Deletion | Chr12 | 16190001 | 16439000 | 249000 | 37 |
| Deletion | Chr12 | 16447001 | 16603000 | 156000 | 20 |
| Deletion | Chr1 | 16642508 | 16642509 | 1 | |
| Deletion | Chr12 | 16651001 | 17111000 | 460000 | 70 |
| Deletion | Chr6 | 16773929 | 16773934 | 5 | |
| Deletion | Chr12 | 17052688 | 17052702 | 14 | |
| Deletion | Chr12 | 22941095 | 22941096 | 1 | |
| Deletion | Chr7 | 24147536 | 24147537 | 1 | |
| Deletion | Chr5 | 24404749 | 24404759 | 10 | LOC_Os05g41720 |
| Deletion | Chr12 | 26242849 | 26242850 | 1 | |
| Deletion | Chr4 | 26457481 | 26457498 | 17 | |
| Deletion | Chr1 | 27775272 | 27775273 | 1 | |
| Deletion | Chr3 | 28320949 | 28320957 | 8 | |
| Deletion | Chr4 | 28687291 | 28687292 | 1 | |
| Deletion | Chr3 | 32369328 | 32369340 | 12 | |
| Deletion | Chr1 | 34283957 | 34283977 | 20 | |
| Deletion | Chr4 | 35078669 | 35078690 | 21 | |
| Deletion | Chr2 | 35231262 | 35231267 | 5 | LOC_Os02g57500 |
| Deletion | Chr1 | 42451317 | 42451321 | 4 | LOC_Os01g73234 |
Insertions: 3
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr11 | 27699077 | 27699078 | 2 | |
| Insertion | Chr2 | 5449446 | 5449448 | 3 | |
| Insertion | Chr5 | 153793 | 153793 | 1 |
Inversions: 3
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr2 | 5772843 | 17587091 | |
| Inversion | Chr3 | 33101708 | 33949285 | |
| Inversion | Chr3 | 33103784 | 33949290 | LOC_Os03g58150 |
Translocations: 1
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 4446676 | Chr3 | 33103827 | LOC_Os03g58150 |
