Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1047-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1047-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 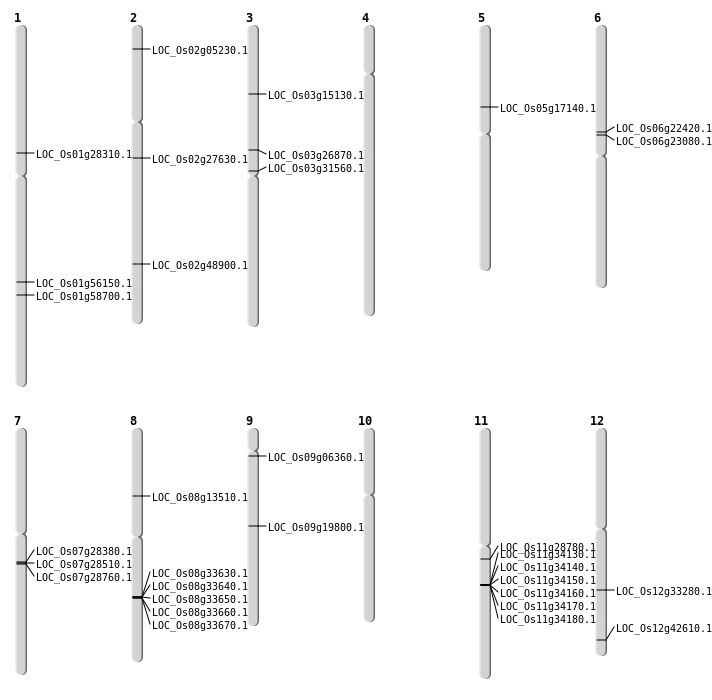
|
| Variant Information |
|---|
Single base substitutions: 35
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 14952885 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 19106605 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 21442377 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 34916803 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 36681689 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 40960773 | A-T | HET | INTERGENIC | |
| SBS | Chr10 | 13152008 | C-G | HET | INTRON | |
| SBS | Chr11 | 16679323 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os11g28780 |
| SBS | Chr11 | 25798806 | C-G | HET | INTERGENIC | |
| SBS | Chr12 | 16330400 | T-G | HOMO | INTRON | |
| SBS | Chr12 | 26479204 | G-A | HET | INTRON | |
| SBS | Chr2 | 34346557 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 35377489 | C-A | HET | INTRON | |
| SBS | Chr2 | 704758 | T-C | HET | INTERGENIC | |
| SBS | Chr2 | 999902 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 15334337 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g26870 |
| SBS | Chr3 | 22779235 | A-C | HET | INTERGENIC | |
| SBS | Chr3 | 9838659 | T-G | HET | INTERGENIC | |
| SBS | Chr4 | 10724103 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 22887598 | G-A | HOMO | INTRON | |
| SBS | Chr4 | 22887599 | T-A | HOMO | INTRON | |
| SBS | Chr4 | 22887600 | G-A | HOMO | INTRON | |
| SBS | Chr4 | 6942766 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 15483154 | T-A | HET | INTRON | |
| SBS | Chr5 | 21856773 | C-A | HOMO | INTERGENIC | |
| SBS | Chr5 | 23816746 | C-A | HOMO | INTERGENIC | |
| SBS | Chr5 | 9810536 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os05g17140 |
| SBS | Chr6 | 28962474 | A-G | HOMO | INTERGENIC | |
| SBS | Chr7 | 16590654 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os07g28380 |
| SBS | Chr7 | 16835086 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os07g28760 |
| SBS | Chr7 | 20163930 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 26040846 | C-T | HOMO | INTRON | |
| SBS | Chr8 | 8036405 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os08g13510 |
| SBS | Chr9 | 22642418 | G-T | HET | INTERGENIC | |
| SBS | Chr9 | 2992485 | C-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os09g06360 |
Deletions: 26
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr2 | 2506377 | 2506406 | 29 | LOC_Os02g05230 |
| Deletion | Chr10 | 3178522 | 3178526 | 4 | |
| Deletion | Chr3 | 5463260 | 5463262 | 2 | |
| Deletion | Chr3 | 7522766 | 7522768 | 2 | |
| Deletion | Chr9 | 8076120 | 8076128 | 8 | |
| Deletion | Chr1 | 10210028 | 10210030 | 2 | |
| Deletion | Chr9 | 11207701 | 11207702 | 1 | |
| Deletion | Chr9 | 11853148 | 11853149 | 1 | LOC_Os09g19800 |
| Deletion | Chr1 | 13009740 | 13009754 | 14 | |
| Deletion | Chr6 | 13032013 | 13032015 | 2 | LOC_Os06g22420 |
| Deletion | Chr1 | 15857318 | 15860947 | 3629 | LOC_Os01g28310 |
| Deletion | Chr7 | 16687867 | 16687877 | 10 | LOC_Os07g28510 |
| Deletion | Chr3 | 18009196 | 18009221 | 25 | LOC_Os03g31560 |
| Deletion | Chr4 | 19438046 | 19438058 | 12 | |
| Deletion | Chr11 | 19987001 | 20009000 | 22000 | 6 |
| Deletion | Chr8 | 21003223 | 21030066 | 26843 | 5 |
| Deletion | Chr8 | 22010324 | 22010330 | 6 | |
| Deletion | Chr5 | 22360554 | 22360556 | 2 | |
| Deletion | Chr4 | 29128072 | 29128081 | 9 | |
| Deletion | Chr2 | 29914390 | 29914394 | 4 | LOC_Os02g48900 |
| Deletion | Chr1 | 30499711 | 30499716 | 5 | |
| Deletion | Chr2 | 31939175 | 31939176 | 1 | |
| Deletion | Chr3 | 33262209 | 33262226 | 17 | |
| Deletion | Chr1 | 33931070 | 33931085 | 15 | LOC_Os01g58700 |
| Deletion | Chr2 | 34784407 | 34784408 | 1 | |
| Deletion | Chr1 | 36752642 | 36752657 | 15 |
Insertions: 1
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr4 | 33899640 | 33899640 | 1 |
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr7 | 7194114 | 8432162 |
Translocations: 6
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr6 | 13456063 | Chr2 | 16375780 | 2 |
| Translocation | Chr4 | 16416709 | Chr1 | 32335247 | LOC_Os01g56150 |
| Translocation | Chr11 | 20126825 | Chr3 | 8265500 | LOC_Os03g15130 |
| Translocation | Chr12 | 20138349 | Chr5 | 13277466 | LOC_Os12g33280 |
| Translocation | Chr12 | 26422802 | Chr1 | 38696850 | |
| Translocation | Chr12 | 26479258 | Chr1 | 38696806 | LOC_Os12g42610 |
