Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1058-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1058-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 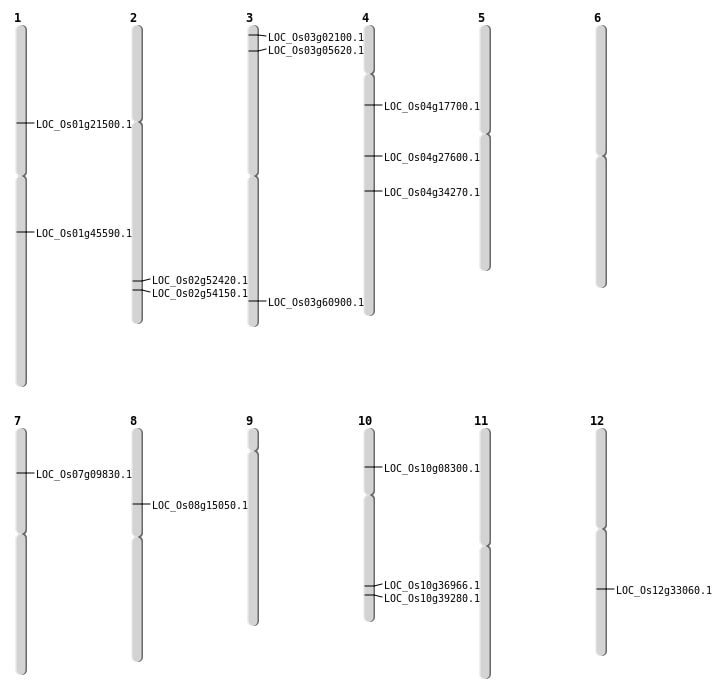
|
| Variant Information |
|---|
Single base substitutions: 45
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 23013105 | A-T | HET | INTERGENIC | |
| SBS | Chr1 | 2552537 | A-G | HOMO | INTERGENIC | |
| SBS | Chr1 | 26418162 | G-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 39063355 | C-T | HOMO | INTERGENIC | |
| SBS | Chr10 | 19854108 | G-A | HET | UTR_3_PRIME | |
| SBS | Chr10 | 4484718 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os10g08300 |
| SBS | Chr11 | 1161992 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 15135673 | C-T | HOMO | INTERGENIC | |
| SBS | Chr11 | 22361247 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 25659838 | G-A | HOMO | INTRON | |
| SBS | Chr12 | 13705825 | C-T | HET | INTRON | |
| SBS | Chr12 | 19978893 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os12g33060 |
| SBS | Chr12 | 21365985 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 23514800 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 25974203 | C-T | HET | INTRON | |
| SBS | Chr2 | 11727838 | C-A | HET | INTRON | |
| SBS | Chr2 | 15800349 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 26097231 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 35385192 | A-G | HET | INTERGENIC | |
| SBS | Chr3 | 18863352 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 6900466 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 16154 | T-A | HET | INTERGENIC | |
| SBS | Chr4 | 16323011 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g27600 |
| SBS | Chr4 | 19542456 | C-A | HET | INTERGENIC | |
| SBS | Chr4 | 20760125 | A-G | HOMO | NON_SYNONYMOUS_CODING | LOC_Os04g34270 |
| SBS | Chr4 | 22861866 | T-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 9703297 | T-G | HET | NON_SYNONYMOUS_CODING | LOC_Os04g17700 |
| SBS | Chr5 | 17872102 | C-T | HET | INTRON | |
| SBS | Chr5 | 3973788 | G-C | HET | INTERGENIC | |
| SBS | Chr6 | 17364850 | G-A | HOMO | INTERGENIC | |
| SBS | Chr6 | 22059152 | T-C | HET | INTERGENIC | |
| SBS | Chr7 | 18932209 | C-T | HOMO | UTR_3_PRIME | |
| SBS | Chr7 | 4969099 | T-A | HET | INTERGENIC | |
| SBS | Chr7 | 5230408 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os07g09830 |
| SBS | Chr7 | 8135698 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 17525704 | A-T | HET | INTERGENIC | |
| SBS | Chr8 | 27746761 | C-T | HOMO | INTERGENIC | |
| SBS | Chr8 | 3269604 | T-A | HOMO | INTERGENIC | |
| SBS | Chr8 | 476592 | G-T | HET | INTRON | |
| SBS | Chr8 | 9098972 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os08g15050 |
| SBS | Chr8 | 9098973 | C-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os08g15050 |
| SBS | Chr9 | 21069772 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 21154711 | G-C | HET | INTERGENIC | |
| SBS | Chr9 | 2208603 | T-C | HOMO | INTRON | |
| SBS | Chr9 | 3162117 | G-A | HOMO | INTRON |
Deletions: 22
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr3 | 674055 | 674056 | 1 | LOC_Os03g02100 |
| Deletion | Chr1 | 727051 | 727063 | 12 | |
| Deletion | Chr3 | 2808753 | 2808758 | 5 | LOC_Os03g05620 |
| Deletion | Chr5 | 5463505 | 5463514 | 9 | |
| Deletion | Chr2 | 8593162 | 8593165 | 3 | |
| Deletion | Chr8 | 10860345 | 10860346 | 1 | |
| Deletion | Chr2 | 11727820 | 11727834 | 14 | |
| Deletion | Chr1 | 12011413 | 12011425 | 12 | LOC_Os01g21500 |
| Deletion | Chr2 | 13839713 | 13839717 | 4 | |
| Deletion | Chr7 | 14153146 | 14153149 | 3 | |
| Deletion | Chr10 | 14291849 | 14291854 | 5 | |
| Deletion | Chr10 | 14529003 | 14529005 | 2 | |
| Deletion | Chr7 | 18022398 | 18022399 | 1 | |
| Deletion | Chr11 | 21310503 | 21310506 | 3 | |
| Deletion | Chr4 | 23346072 | 23346079 | 7 | |
| Deletion | Chr1 | 25876923 | 25876925 | 2 | |
| Deletion | Chr1 | 25893666 | 25893672 | 6 | LOC_Os01g45590 |
| Deletion | Chr2 | 32081424 | 32081439 | 15 | LOC_Os02g52420 |
| Deletion | Chr2 | 33192443 | 33192444 | 1 | LOC_Os02g54150 |
| Deletion | Chr2 | 33923199 | 33923224 | 25 | |
| Deletion | Chr3 | 34607813 | 34608996 | 1183 | LOC_Os03g60900 |
| Deletion | Chr1 | 39610479 | 39610520 | 41 |
Insertions: 3
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 7964968 | 7964968 | 1 | |
| Insertion | Chr10 | 2738516 | 2738531 | 16 | |
| Insertion | Chr11 | 12653359 | 12653360 | 2 |
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr10 | 19807776 | 20965629 | 2 |
No Translocation
