Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1062-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1062-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 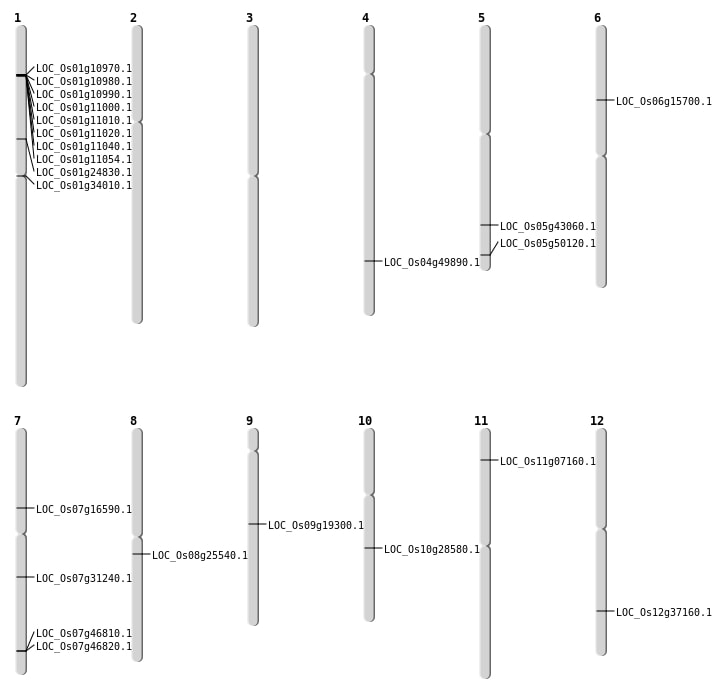
|
| Variant Information |
|---|
Single base substitutions: 39
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 13985196 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g24830 |
| SBS | Chr1 | 14134856 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 16702561 | T-G | HET | INTERGENIC | |
| SBS | Chr1 | 18722266 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os01g34010 |
| SBS | Chr1 | 21148845 | T-C | HET | INTERGENIC | |
| SBS | Chr1 | 2271608 | G-T | HET | SPLICE_SITE_REGION | |
| SBS | Chr1 | 24266153 | C-T | HET | INTRON | |
| SBS | Chr1 | 39495897 | G-A | HOMO | INTRON | |
| SBS | Chr10 | 975169 | A-T | HET | INTERGENIC | |
| SBS | Chr11 | 24752943 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 12632873 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr12 | 1368173 | G-A | HOMO | INTERGENIC | |
| SBS | Chr12 | 24088446 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr12 | 9188352 | T-G | HOMO | INTERGENIC | |
| SBS | Chr2 | 20192939 | T-A | HET | INTERGENIC | |
| SBS | Chr2 | 335363 | A-G | HET | INTERGENIC | |
| SBS | Chr2 | 33765168 | T-C | HOMO | INTERGENIC | |
| SBS | Chr3 | 17880026 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 35386450 | T-G | HOMO | INTRON | |
| SBS | Chr3 | 6271570 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 10670140 | G-T | HET | INTRON | |
| SBS | Chr5 | 17982400 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 23100668 | T-G | HET | INTERGENIC | |
| SBS | Chr5 | 26784993 | T-A | HET | INTERGENIC | |
| SBS | Chr5 | 26784994 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 12606826 | G-C | HET | INTERGENIC | |
| SBS | Chr6 | 23881947 | C-G | HET | INTERGENIC | |
| SBS | Chr6 | 7277619 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 7600379 | G-T | HET | INTERGENIC | |
| SBS | Chr7 | 15958490 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 18491250 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os07g31240 |
| SBS | Chr7 | 3245957 | G-T | HOMO | INTRON | |
| SBS | Chr7 | 9731159 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os07g16590 |
| SBS | Chr8 | 15534491 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os08g25540 |
| SBS | Chr8 | 16926185 | T-A | HET | INTERGENIC | |
| SBS | Chr9 | 11551751 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os09g19300 |
| SBS | Chr9 | 19234959 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 21427272 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 9172648 | T-C | HET | INTERGENIC |
Deletions: 28
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr2 | 1737241 | 1737248 | 7 | |
| Deletion | Chr9 | 1861642 | 1861654 | 12 | |
| Deletion | Chr7 | 2149402 | 2149403 | 1 | |
| Deletion | Chr11 | 3568460 | 3568485 | 25 | LOC_Os11g07160 |
| Deletion | Chr6 | 3811035 | 3811038 | 3 | |
| Deletion | Chr6 | 5029349 | 5029351 | 2 | |
| Deletion | Chr1 | 5844001 | 5907000 | 63000 | 8 |
| Deletion | Chr5 | 5951381 | 5951394 | 13 | |
| Deletion | Chr7 | 5956597 | 5956601 | 4 | |
| Deletion | Chr10 | 6502968 | 6502969 | 1 | |
| Deletion | Chr7 | 6755146 | 6755147 | 1 | |
| Deletion | Chr8 | 6766584 | 6766585 | 1 | |
| Deletion | Chr6 | 8892556 | 8892567 | 11 | LOC_Os06g15700 |
| Deletion | Chr4 | 10248648 | 10248649 | 1 | |
| Deletion | Chr10 | 13796321 | 13796338 | 17 | |
| Deletion | Chr9 | 14829726 | 14829728 | 2 | |
| Deletion | Chr10 | 14874911 | 14874914 | 3 | LOC_Os10g28580 |
| Deletion | Chr4 | 18609607 | 18609612 | 5 | |
| Deletion | Chr12 | 19256137 | 19256146 | 9 | |
| Deletion | Chr12 | 20076156 | 20076206 | 50 | |
| Deletion | Chr4 | 20451855 | 20451857 | 2 | |
| Deletion | Chr12 | 22785107 | 22785108 | 1 | LOC_Os12g37160 |
| Deletion | Chr5 | 25013809 | 25013863 | 54 | LOC_Os05g43060 |
| Deletion | Chr12 | 25346895 | 25346896 | 1 | |
| Deletion | Chr1 | 26881449 | 26881450 | 1 | |
| Deletion | Chr7 | 27981173 | 27983823 | 2650 | 2 |
| Deletion | Chr5 | 28722351 | 28722352 | 1 | LOC_Os05g50120 |
| Deletion | Chr4 | 29744231 | 29744232 | 1 | LOC_Os04g49890 |
Insertions: 3
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 41613118 | 41613119 | 2 | |
| Insertion | Chr11 | 9774565 | 9774565 | 1 | |
| Insertion | Chr7 | 13222928 | 13222928 | 1 |
No Inversion
No Translocation
