Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1065-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1065-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 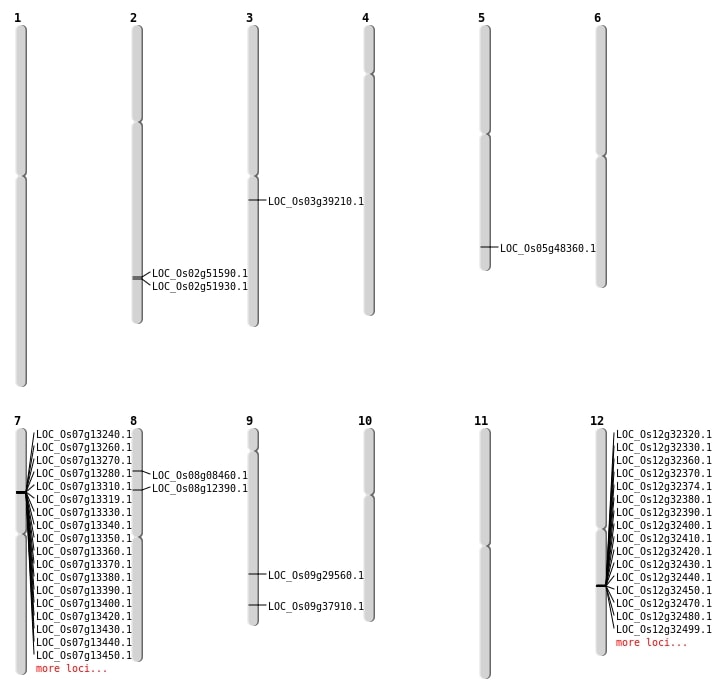
|
| Variant Information |
|---|
Single base substitutions: 46
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 103919 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 20173167 | C-T | HOMO | INTRON | |
| SBS | Chr1 | 38517452 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 21470423 | G-T | HOMO | INTRON | |
| SBS | Chr11 | 19580986 | A-T | HOMO | INTRON | |
| SBS | Chr11 | 22240200 | T-C | HOMO | INTERGENIC | |
| SBS | Chr12 | 1773162 | C-T | HET | UTR_3_PRIME | |
| SBS | Chr12 | 18016459 | T-C | HET | INTERGENIC | |
| SBS | Chr12 | 18317899 | T-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr2 | 10316559 | T-C | HET | INTERGENIC | |
| SBS | Chr2 | 8132699 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 85362 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr3 | 10023228 | T-G | HOMO | INTERGENIC | |
| SBS | Chr3 | 21775543 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os03g39210 |
| SBS | Chr3 | 725730 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 9744946 | A-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr4 | 27689346 | A-G | HET | INTERGENIC | |
| SBS | Chr4 | 3114322 | T-A | HET | INTERGENIC | |
| SBS | Chr5 | 17127002 | A-G | HET | INTERGENIC | |
| SBS | Chr5 | 27726670 | C-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os05g48360 |
| SBS | Chr5 | 28531703 | C-A | HET | INTERGENIC | |
| SBS | Chr5 | 7927721 | C-T | HET | INTRON | |
| SBS | Chr5 | 8611978 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 11350818 | C-T | HOMO | INTERGENIC | |
| SBS | Chr6 | 13134429 | G-A | HOMO | INTERGENIC | |
| SBS | Chr6 | 24298495 | A-G | HET | INTRON | |
| SBS | Chr6 | 24606904 | T-A | HOMO | INTERGENIC | |
| SBS | Chr6 | 26502095 | C-T | HOMO | INTERGENIC | |
| SBS | Chr6 | 28107316 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 10832288 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 14002345 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 14272145 | T-C | HET | INTERGENIC | |
| SBS | Chr7 | 22850034 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 23501897 | A-G | HET | INTERGENIC | |
| SBS | Chr7 | 26900687 | G-T | HET | INTERGENIC | |
| SBS | Chr7 | 5905056 | A-C | HET | INTERGENIC | |
| SBS | Chr7 | 5905057 | C-A | HET | INTERGENIC | |
| SBS | Chr7 | 9144649 | A-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr8 | 11099052 | G-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr8 | 12733247 | C-A | HOMO | INTERGENIC | |
| SBS | Chr8 | 26600152 | G-A | HOMO | INTERGENIC | |
| SBS | Chr8 | 4872092 | A-G | HOMO | NON_SYNONYMOUS_CODING | LOC_Os08g08460 |
| SBS | Chr8 | 7311360 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os08g12390 |
| SBS | Chr9 | 10886797 | C-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr9 | 12687142 | A-G | HET | INTERGENIC | |
| SBS | Chr9 | 17978389 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os09g29560 |
Deletions: 17
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr7 | 7590001 | 8247000 | 657000 | 114 |
| Deletion | Chr11 | 8444609 | 8444611 | 2 | |
| Deletion | Chr11 | 10547129 | 10547134 | 5 | |
| Deletion | Chr11 | 11618001 | 11618033 | 32 | |
| Deletion | Chr11 | 12013294 | 12013295 | 1 | |
| Deletion | Chr10 | 17497114 | 17497131 | 17 | |
| Deletion | Chr6 | 17730275 | 17730280 | 5 | |
| Deletion | Chr11 | 18693261 | 18693264 | 3 | |
| Deletion | Chr12 | 19507001 | 20159000 | 652000 | 93 |
| Deletion | Chr10 | 20153841 | 20153848 | 7 | |
| Deletion | Chr12 | 20219001 | 20428000 | 209000 | 40 |
| Deletion | Chr7 | 23880312 | 23880353 | 41 | |
| Deletion | Chr12 | 23936322 | 23936330 | 8 | LOC_Os12g38930 |
| Deletion | Chr2 | 29470256 | 29470262 | 6 | |
| Deletion | Chr2 | 31600535 | 31600616 | 81 | LOC_Os02g51590 |
| Deletion | Chr2 | 31807418 | 31807435 | 17 | LOC_Os02g51930 |
| Deletion | Chr2 | 35681272 | 35681279 | 7 |
Insertions: 5
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr9 | 21614253 | 21857080 | LOC_Os09g37910 |
| Inversion | Chr9 | 21857094 | 22707294 | LOC_Os09g37910 |
Translocations: 7
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr8 | 2594411 | Chr1 | 17033505 | |
| Translocation | Chr8 | 4022945 | Chr6 | 6194036 | |
| Translocation | Chr8 | 4023195 | Chr7 | 7588570 | |
| Translocation | Chr7 | 7588571 | Chr6 | 6194036 | |
| Translocation | Chr7 | 7589227 | Chr6 | 6194029 | |
| Translocation | Chr9 | 21857094 | Chr3 | 9468158 | LOC_Os09g37910 |
| Translocation | Chr9 | 22707294 | Chr3 | 9468443 |
